
| Name: MAP6D1 | Sequence: fasta or formatted (199aa) | NCBI GI: 13376306 | |
|
Description: MAP6 domain containing 1
|
Referenced in:
| ||
|
Composition:
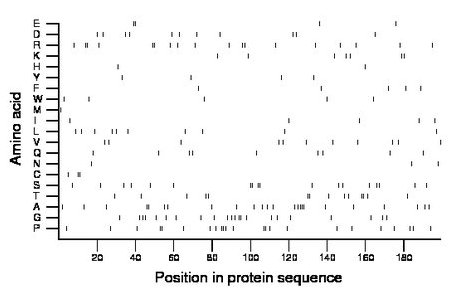
Amino acid Percentage Count Longest homopolymer A alanine 12.1 24 4 C cysteine 1.5 3 2 D aspartate 5.5 11 1 E glutamate 2.0 4 2 F phenylalanine 2.5 5 1 G glycine 11.1 22 2 H histidine 1.0 2 1 I isoleucine 2.5 5 1 K lysine 3.5 7 2 L leucine 5.0 10 1 M methionine 0.5 1 1 N asparagine 1.5 3 1 P proline 11.6 23 3 Q glutamine 4.5 9 1 R arginine 9.0 18 2 S serine 9.0 18 2 T threonine 7.0 14 2 V valine 5.5 11 1 W tryptophan 2.5 5 1 Y tyrosine 2.0 4 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 MAP6 domain containing 1 MAP6 0.133 microtubule-associated protein 6 isoform 2 MAP6 0.133 microtubule-associated protein 6 isoform 1 HNRNPK 0.038 heterogeneous nuclear ribonucleoprotein K isoform a ... HNRNPK 0.038 heterogeneous nuclear ribonucleoprotein K isoform b ... HNRNPK 0.038 heterogeneous nuclear ribonucleoprotein K isoform a ... LOC100131851 0.031 PREDICTED: hypothetical protein LOC100130751 0.031 PREDICTED: hypothetical protein KIAA0907 0.031 hypothetical protein LOC22889 ZCCHC3 0.031 zinc finger, CCHC domain containing 3 WBP2NL 0.028 WBP2 N-terminal like COL1A1 0.028 alpha 1 type I collagen preproprotein LOC100131851 0.026 PREDICTED: hypothetical protein XYLT1 0.026 xylosyltransferase I OGFRL1 0.026 opioid growth factor receptor-like 1 SFTPD 0.026 pulmonary surfactant-associated protein D precursor ... STAP2 0.026 signal transducing adaptor family member 2 isoform 2... STAP2 0.026 signal transducing adaptor family member 2 isoform 1... LOC100293375 0.026 PREDICTED: hypothetical protein FMNL3 0.026 formin-like 3 isoform 2 FMNL3 0.026 formin-like 3 isoform 1 KIAA0649 0.026 1A6/DRIM (down-regulated in metastasis) interacting ... COL12A1 0.023 collagen, type XII, alpha 1 short isoform precursor ... COL12A1 0.023 collagen, type XII, alpha 1 long isoform precursor [... CNOT3 0.023 CCR4-NOT transcription complex, subunit 3 COL23A1 0.023 collagen, type XXIII, alpha 1 WNK4 0.020 WNK lysine deficient protein kinase 4 LOC100292058 0.020 PREDICTED: hypothetical protein XP_002344766 LOC100130503 0.020 PREDICTED: hypothetical protein LOC645321 0.020 PREDICTED: hypothetical proteinHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
