
| Name: N4BP1 | Sequence: fasta or formatted (896aa) | NCBI GI: 48928019 | |
|
Description: Nedd4 binding protein 1
|
Referenced in:
| ||
|
Composition:
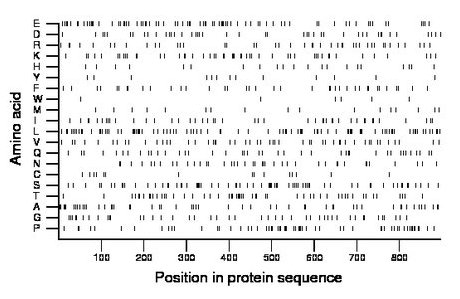
Amino acid Percentage Count Longest homopolymer A alanine 6.0 54 2 C cysteine 1.8 16 2 D aspartate 5.0 45 2 E glutamate 9.2 82 2 F phenylalanine 4.4 39 2 G glycine 4.9 44 2 H histidine 2.3 21 1 I isoleucine 4.9 44 2 K lysine 5.6 50 2 L leucine 10.5 94 4 M methionine 2.0 18 1 N asparagine 4.6 41 2 P proline 6.8 61 2 Q glutamine 5.2 47 2 R arginine 5.0 45 2 S serine 7.9 71 3 T threonine 6.0 54 2 V valine 5.7 51 2 W tryptophan 0.7 6 1 Y tyrosine 1.5 13 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 Nedd4 binding protein 1 KIAA0323 0.128 hypothetical protein LOC23351 KIAA1305 0.101 hypothetical protein LOC57523 ZC3H12C 0.085 zinc finger CCCH-type containing 12C ZC3H12A 0.084 zinc finger CCCH-type containing 12A ZC3H12B 0.083 zinc finger CCCH-type containing 12B ZC3H12D 0.077 zinc finger CCCH-type containing 12D C12orf35 0.008 hypothetical protein LOC55196 CACNA2D2 0.008 calcium channel, voltage-dependent, alpha 2/delta su... CACNA2D2 0.008 calcium channel, voltage-dependent, alpha 2/delta su... LOC728503 0.008 PREDICTED: hypothetical protein LOC728503 0.008 PREDICTED: hypothetical protein LOC728503 0.008 PREDICTED: hypothetical protein CEP63 0.008 centrosomal protein 63 isoform a MYH10 0.007 myosin, heavy polypeptide 10, non-muscle PHF14 0.007 PHD finger protein 14 isoform 2 PHF14 0.007 PHD finger protein 14 isoform 1 GOLGB1 0.007 golgi autoantigen, golgin subfamily b, macrogolgin ... CENPF 0.006 centromere protein F MTUS1 0.006 mitochondrial tumor suppressor 1 isoform 5 MTUS1 0.006 mitochondrial tumor suppressor 1 isoform 1 MTUS1 0.006 mitochondrial tumor suppressor 1 isoform 2 MTUS1 0.006 mitochondrial tumor suppressor 1 isoform 4 EEA1 0.006 early endosome antigen 1, 162kD CGNL1 0.006 cingulin-like 1 POM121C 0.006 POM121 membrane glycoprotein (rat)-like PRL 0.006 prolactin precursor MLL5 0.006 myeloid/lymphoid or mixed-lineage leukemia 5 MLL5 0.006 myeloid/lymphoid or mixed-lineage leukemia 5 SAP130 0.006 Sin3A-associated protein, 130kDa isoform bHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
