
| Name: CD3E | Sequence: fasta or formatted (207aa) | NCBI GI: 4502671 | |
|
Description: CD3E antigen, epsilon polypeptide precursor
|
Referenced in:
| ||
|
Composition:
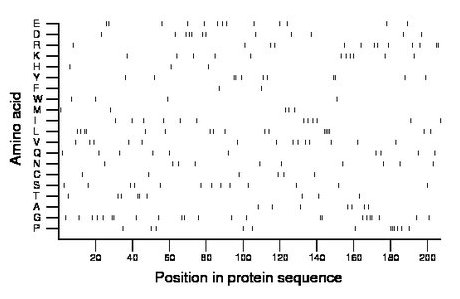
Amino acid Percentage Count Longest homopolymer A alanine 3.4 7 1 C cysteine 2.9 6 1 D aspartate 5.8 12 2 E glutamate 6.3 13 2 F phenylalanine 1.0 2 1 G glycine 11.1 23 2 H histidine 1.4 3 1 I isoleucine 5.8 12 1 K lysine 5.3 11 1 L leucine 8.2 17 4 M methionine 2.4 5 1 N asparagine 4.8 10 1 P proline 5.8 12 3 Q glutamine 4.8 10 1 R arginine 6.3 13 2 S serine 6.3 13 1 T threonine 4.3 9 2 V valine 6.8 14 1 W tryptophan 1.9 4 1 Y tyrosine 5.3 11 2 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 CD3E antigen, epsilon polypeptide precursor CD3G 0.046 CD3G antigen, gamma polypeptide precursor PXDNL 0.036 peroxidasin homolog-like UNC5D 0.019 unc-5 homolog D SCN4B 0.017 sodium channel, voltage-gated, type IV, beta isoform... SCN4B 0.017 sodium channel, voltage-gated, type IV, beta isofor... CD3D 0.017 CD3 antigen, delta subunit isoform A precursor AHNAK 0.012 AHNAK nucleoprotein isoform 1 NDST2 0.012 heparan glucosaminyl N-deacetylase/N-sulfotransferase... CABIN1 0.010 calcineurin binding protein 1 TTN 0.010 titin isoform novex-2 TTN 0.010 titin isoform novex-1 TTN 0.010 titin isoform N2-B KIR2DS1 0.010 killer cell immunoglobulin-like receptor, two domains... TSPAN6 0.010 transmembrane 4 superfamily member 6 BRWD3 0.010 bromodomain and WD repeat domain containing 3 [Homo... KIR2DS3 0.007 killer cell immunoglobulin-like receptor, two domains... MAP1A 0.007 microtubule-associated protein 1A SEMA3C 0.007 semaphorin 3C PHF14 0.007 PHD finger protein 14 isoform 2 PHF14 0.007 PHD finger protein 14 isoform 1 TTN 0.007 titin isoform N2-A PRKAR1A 0.007 cAMP-dependent protein kinase, regulatory subunit al... PRKAR1A 0.007 cAMP-dependent protein kinase, regulatory subunit al... PRKAR1A 0.007 cAMP-dependent protein kinase, regulatory subunit alp... KIR2DL1 0.007 killer cell immunoglobulin-like receptor, two domai... BOC 0.007 brother of CDO DRGX 0.007 dorsal root ganglia homeobox LOC100289869 0.007 PREDICTED: hypothetical protein XP_002347023 LOC100291142 0.007 PREDICTED: hypothetical protein XP_002347004Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
