
| Name: BICC1 | Sequence: fasta or formatted (974aa) | NCBI GI: 122937472 | |
|
Description: bicaudal C homolog 1
|
Referenced in: Additional Genes in Development
| ||
|
Composition:
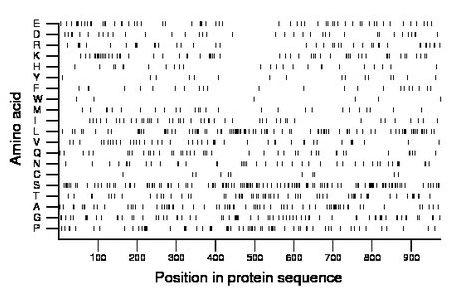
Amino acid Percentage Count Longest homopolymer A alanine 7.8 76 4 C cysteine 1.1 11 1 D aspartate 3.7 36 2 E glutamate 5.9 57 2 F phenylalanine 2.4 23 2 G glycine 7.4 72 3 H histidine 2.8 27 2 I isoleucine 4.6 45 1 K lysine 6.3 61 2 L leucine 9.2 90 2 M methionine 3.1 30 2 N asparagine 4.6 45 2 P proline 6.6 64 3 Q glutamine 4.5 44 3 R arginine 4.2 41 2 S serine 12.6 123 3 T threonine 6.4 62 3 V valine 4.6 45 2 W tryptophan 0.6 6 1 Y tyrosine 1.6 16 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 bicaudal C homolog 1 ANKS6 0.028 ankyrin repeat and sterile alpha motif domain contai... HDLBP 0.027 high density lipoprotein binding protein HDLBP 0.027 high density lipoprotein binding protein PCBP2 0.023 poly(rC) binding protein 2 isoform c ANKS3 0.022 ankyrin repeat and sterile alpha motif domain contai... PCBP2 0.021 poly(rC) binding protein 2 isoform f PCBP2 0.021 poly(rC) binding protein 2 isoform g PCBP2 0.017 poly(rC) binding protein 2 isoform b PCBP3 0.017 poly(rC) binding protein 3 isoform 1 PCBP3 0.016 poly(rC) binding protein 3 isoform 2 PCBP1 0.015 poly(rC) binding protein 1 PCBP2 0.015 poly(rC) binding protein 2 isoform a PCBP2 0.015 poly(rC) binding protein 2 isoform e PCBP4 0.015 poly(rC) binding protein 4 isoform c PCBP4 0.015 poly(rC) binding protein 4 isoform c PCBP2 0.013 poly(rC) binding protein 2 isoform d MUC5AC 0.012 mucin 5AC TNKS2 0.012 tankyrase, TRF1-interacting ankyrin-related ADP-ribo... SPATC1 0.011 spermatogenesis and centriole associated 1 isoform ... SPATC1 0.011 spermatogenesis and centriole associated 1 isoform ... SRCAP 0.011 Snf2-related CBP activator protein MUC5B 0.011 mucin 5, subtype B, tracheobronchial MUC17 0.011 mucin 17 ZCCHC14 0.010 zinc finger, CCHC domain containing 14 DUSP27 0.009 dual specificity phosphatase 27 NCAN 0.009 chondroitin sulfate proteoglycan 3 MEX3A 0.009 MEX3A protein IGF2BP2 0.009 insulin-like growth factor 2 mRNA binding protein 2 ... MUC12 0.009 PREDICTED: mucin 12Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
