
| Name: FAM9C | Sequence: fasta or formatted (166aa) | NCBI GI: 28274703 | |
|
Description: family with sequence similarity 9, member C
|
Referenced in:
| ||
|
Composition:
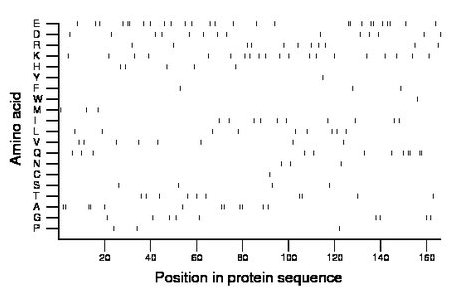
Amino acid Percentage Count Longest homopolymer A alanine 7.2 12 2 C cysteine 0.6 1 1 D aspartate 8.4 14 1 E glutamate 16.3 27 2 F phenylalanine 1.8 3 1 G glycine 5.4 9 1 H histidine 3.0 5 1 I isoleucine 6.0 10 1 K lysine 12.0 20 1 L leucine 5.4 9 1 M methionine 1.8 3 1 N asparagine 1.8 3 1 P proline 1.8 3 1 Q glutamine 7.2 12 2 R arginine 6.6 11 1 S serine 2.4 4 1 T threonine 6.0 10 2 V valine 4.8 8 1 W tryptophan 0.6 1 1 Y tyrosine 0.6 1 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 family with sequence similarity 9, member C FAM9B 0.484 family with sequence similarity 9, member B FAM9A 0.415 family with sequence similarity 9, member A SYCP3 0.131 synaptonemal complex protein 3 LOC100133758 0.052 PREDICTED: hypothetical protein, partial CALD1 0.049 caldesmon 1 isoform 1 EIF5B 0.046 eukaryotic translation initiation factor 5B KIF21A 0.042 kinesin family member 21A PAF1 0.042 Paf1, RNA polymerase II associated factor, homolog [... FAM161B 0.039 hypothetical protein LOC145483 PKD2 0.039 polycystin 2 FHAD1 0.036 forkhead-associated (FHA) phosphopeptide binding do... INCENP 0.036 inner centromere protein antigens 135/155kDa isofor... INCENP 0.036 inner centromere protein antigens 135/155kDa isofor... TCHH 0.033 trichohyalin MAP7D2 0.033 MAP7 domain containing 2 TNNT3 0.033 troponin T3, skeletal, fast isoform 1 TRIM38 0.033 tripartite motif-containing 38 KRT23 0.029 keratin 23 LOC400352 0.029 PREDICTED: similar to Putative golgin subfamily A m... LOC100132816 0.029 PREDICTED: similar to Putative golgin subfamily A m... RPGR 0.029 retinitis pigmentosa GTPase regulator isoform C [Hom... SH2D4B 0.029 SH2 domain containing 4B isoform 2 SH2D4B 0.029 SH2 domain containing 4B isoform 1 GAS7 0.026 growth arrest-specific 7 isoform d GAS7 0.026 growth arrest-specific 7 isoform c GAS7 0.026 growth arrest-specific 7 isoform b GAS7 0.026 growth arrest-specific 7 isoform a SMC2 0.026 structural maintenance of chromosomes 2 SMC2 0.026 structural maintenance of chromosomes 2Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
