
| Name: IER3 | Sequence: fasta or formatted (156aa) | NCBI GI: 119964723 | |
|
Description: immediate early response 3
|
Referenced in: Additional Stress Response Functions
| ||
|
Composition:
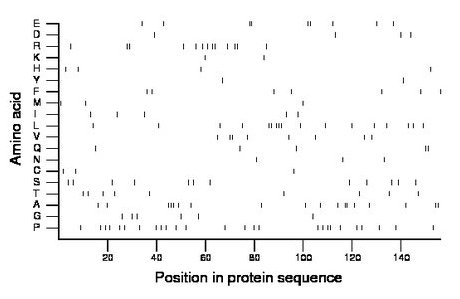
Amino acid Percentage Count Longest homopolymer A alanine 11.5 18 3 C cysteine 1.9 3 1 D aspartate 2.6 4 1 E glutamate 5.8 9 2 F phenylalanine 4.5 7 1 G glycine 3.8 6 1 H histidine 2.6 4 1 I isoleucine 3.2 5 1 K lysine 1.3 2 1 L leucine 10.9 17 3 M methionine 1.9 3 1 N asparagine 1.9 3 1 P proline 16.7 26 2 Q glutamine 3.2 5 2 R arginine 8.3 13 2 S serine 7.7 12 1 T threonine 5.8 9 1 V valine 5.1 8 2 W tryptophan 0.0 0 0 Y tyrosine 1.3 2 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 immediate early response 3 C9orf167 0.081 hypothetical protein LOC54863 SRCAP 0.054 Snf2-related CBP activator protein KAT2A 0.047 general control of amino acid synthesis 5-like 2 [H... SETD1A 0.047 SET domain containing 1A LOC729580 0.041 PREDICTED: hypothetical protein LOC729580 0.041 PREDICTED: hypothetical protein LOC729580 0.041 PREDICTED: hypothetical protein DGKK 0.041 diacylglycerol kinase kappa BCORL1 0.041 BCL6 co-repressor-like 1 KIAA0284 0.041 hypothetical protein LOC283638 isoform 2 KIAA0284 0.041 hypothetical protein LOC283638 isoform 1 LOC400236 0.037 PREDICTED: hypothetical protein LOC400236 0.037 PREDICTED: hypothetical protein ANKRD33B 0.037 PREDICTED: ankyrin repeat domain 33B UBE2O 0.037 ubiquitin-conjugating enzyme E2O TET3 0.034 tet oncogene family member 3 C10orf95 0.034 hypothetical protein LOC79946 GRIN2C 0.034 N-methyl-D-aspartate receptor subunit 2C precursor [... NACA 0.034 nascent polypeptide-associated complex alpha subuni... TPRX1 0.031 tetra-peptide repeat homeobox LOC100127891 0.027 PREDICTED: similar to hCG2042508 LOC100127891 0.027 PREDICTED: similar to hCG2042508 LOC100127891 0.027 PREDICTED: similar to hCG2042508 DAB2IP 0.027 disabled homolog 2 interacting protein isoform 2 [Ho... DAB2IP 0.027 disabled homolog 2 interacting protein isoform 1 [Ho... ZFPM1 0.027 zinc finger protein, multitype 1 ADRB1 0.027 beta-1-adrenergic receptor TNRC18 0.027 trinucleotide repeat containing 18 USP51 0.027 ubiquitin specific protease 51Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
