
| Name: LOC100292466 | Sequence: fasta or formatted (82aa) | NCBI GI: 239755528 | |
|
Description: PREDICTED: hypothetical protein
| Not currently referenced in the text | ||
|
Composition:
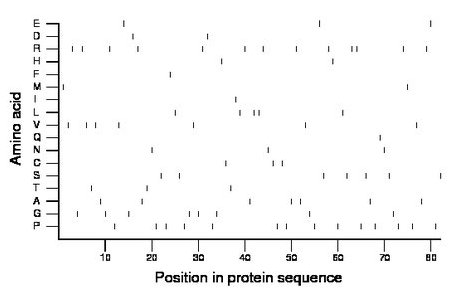
Amino acid Percentage Count Longest homopolymer A alanine 8.5 7 1 C cysteine 3.7 3 1 D aspartate 2.4 2 1 E glutamate 3.7 3 1 F phenylalanine 1.2 1 1 G glycine 9.8 8 1 H histidine 2.4 2 1 I isoleucine 1.2 1 1 K lysine 0.0 0 0 L leucine 6.1 5 2 M methionine 2.4 2 1 N asparagine 3.7 3 1 P proline 17.1 14 1 Q glutamine 1.2 1 1 R arginine 15.9 13 2 S serine 8.5 7 1 T threonine 3.7 3 1 V valine 8.5 7 1 W tryptophan 0.0 0 0 Y tyrosine 0.0 0 0 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein LOC100289816 1.000 PREDICTED: hypothetical protein XP_002347235 LOC100289633 1.000 PREDICTED: hypothetical protein XP_002343096 LOC100292934 0.053 PREDICTED: hypothetical protein LOC100131435 0.047 PREDICTED: hypothetical protein PLXNA4 0.047 plexin A4 isoform 1 NUDT17 0.047 nudix (nucleoside diphosphate linked moiety X)-type ... LOC100128071 0.040 PREDICTED: hypothetical protein LOC100128071 LOC100128071 0.040 PREDICTED: hypothetical protein LOC100128071 LOC100128071 0.040 PREDICTED: hypothetical protein LOC100128071 LOC100131435 0.040 PREDICTED: hypothetical protein LOC100131435 0.040 PREDICTED: hypothetical protein CRH 0.033 corticotropin releasing hormone precursor LOC100131311 0.033 PREDICTED: hypothetical protein LOC100288439 0.033 PREDICTED: hypothetical protein LOC100131311 0.033 PREDICTED: hypothetical protein LOC100131311 0.033 PREDICTED: hypothetical protein LOC100292143 0.033 PREDICTED: hypothetical protein LOC100129103 0.033 PREDICTED: similar to hCG2038970 LOC100132261 0.033 PREDICTED: similar to hCG2023245 SRRM1 0.033 serine/arginine repetitive matrix 1 DMWD 0.027 dystrophia myotonica-containing WD repeat motif prot... LOC730883 0.027 PREDICTED: hypothetical protein LOC730883 0.027 PREDICTED: hypothetical protein SPEG 0.027 SPEG complex locus AFF3 0.020 AF4/FMR2 family, member 3 isoform 2 AFF3 0.020 AF4/FMR2 family, member 3 isoform 1 LOC100292629 0.020 PREDICTED: hypothetical protein LOC100289960 0.020 PREDICTED: hypothetical protein XP_002346853 LOC100286905 0.020 PREDICTED: hypothetical protein XP_002342691Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
