
| Name: LOC100129103 | Sequence: fasta or formatted (915aa) | NCBI GI: 169201839 | |
|
Description: PREDICTED: similar to hCG2038970
| Not currently referenced in the text | ||
Other entries for this name:
alt prot {821aa} PREDICTED: similar to hCG2038970 alt prot {904aa} PREDICTED: similar to hCG2038970 | |||
|
Composition:
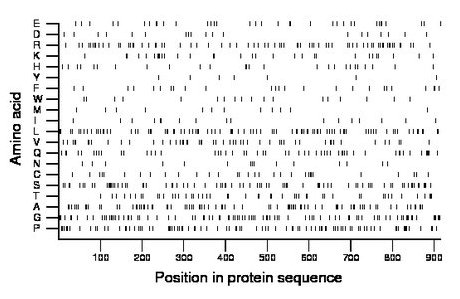
Amino acid Percentage Count Longest homopolymer A alanine 9.6 88 4 C cysteine 3.4 31 1 D aspartate 2.1 19 1 E glutamate 3.9 36 2 F phenylalanine 2.7 25 1 G glycine 9.9 91 3 H histidine 2.8 26 1 I isoleucine 1.9 17 1 K lysine 3.4 31 2 L leucine 10.1 92 3 M methionine 1.4 13 1 N asparagine 1.6 15 1 P proline 10.1 92 3 Q glutamine 5.9 54 2 R arginine 7.9 72 2 S serine 9.3 85 2 T threonine 5.5 50 2 V valine 5.7 52 2 W tryptophan 2.1 19 2 Y tyrosine 0.8 7 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: similar to hCG2038970 LOC100129103 0.827 PREDICTED: similar to hCG2038970 LOC100129103 0.668 PREDICTED: similar to hCG2038970 CXorf67 0.006 hypothetical protein LOC340602 SRRM2 0.006 splicing coactivator subunit SRm300 MUC17 0.006 mucin 17 LOC100291991 0.006 PREDICTED: similar to golgi autoantigen, golgin sub... LOC100290472 0.006 PREDICTED: hypothetical protein XP_002346905 LOC100287825 0.006 PREDICTED: similar to golgi autoantigen, golgin sub... WDR33 0.006 WD repeat domain 33 isoform 1 RERE 0.005 atrophin-1 like protein isoform b RERE 0.005 atrophin-1 like protein isoform a RERE 0.005 atrophin-1 like protein isoform a RADIL 0.005 Rap GTPase interactor CHRD 0.005 chordin LOC100293375 0.005 PREDICTED: hypothetical protein LOC100130311 0.005 hypothetical protein LOC100130311 LOC100287825 0.005 PREDICTED: hypothetical protein XP_002342738 C10orf91 0.005 hypothetical protein LOC170393 MUC12 0.004 PREDICTED: mucin 12 LOC100132249 0.004 PREDICTED: hypothetical protein KIAA1602 0.004 hypothetical protein LOC57701 GRRP1 0.004 glycine/arginine rich protein 1 C17orf63 0.004 hypothetical protein LOC55731 C17orf63 0.004 hypothetical protein LOC55731 GRID2IP 0.004 glutamate receptor, ionotropic, delta 2 (Grid2) int... OBSCN 0.004 obscurin, cytoskeletal calmodulin and titin-interac... BSN 0.004 bassoon protein ETV3 0.004 ets variant gene 3 isoform 1 LOC730130 0.004 hypothetical protein LOC730130Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
