
| Name: LOC100293254 | Sequence: fasta or formatted (344aa) | NCBI GI: 239755509 | |
|
Description: PREDICTED: hypothetical protein
| Not currently referenced in the text | ||
|
Composition:
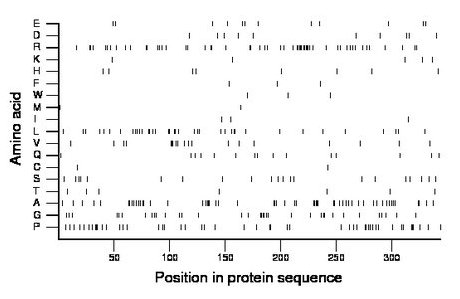
Amino acid Percentage Count Longest homopolymer A alanine 16.3 56 5 C cysteine 0.6 2 1 D aspartate 2.9 10 1 E glutamate 3.5 12 1 F phenylalanine 0.9 3 1 G glycine 10.2 35 4 H histidine 2.3 8 1 I isoleucine 0.9 3 1 K lysine 1.5 5 1 L leucine 11.0 38 2 M methionine 0.6 2 1 N asparagine 0.0 0 0 P proline 14.2 49 3 Q glutamine 4.4 15 1 R arginine 16.3 56 6 S serine 6.1 21 1 T threonine 2.0 7 1 V valine 5.5 19 3 W tryptophan 0.9 3 1 Y tyrosine 0.0 0 0 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein LOC100289377 1.000 PREDICTED: hypothetical protein LOC100289377 1.000 PREDICTED: hypothetical protein XP_002343119 LOC100287104 0.039 PREDICTED: hypothetical protein XP_002343630 LOC100293596 0.036 PREDICTED: similar to mucin LOC100291311 0.036 PREDICTED: hypothetical protein XP_002347439 LOC100289410 0.036 PREDICTED: hypothetical protein XP_002344274 APC2 0.033 adenomatosis polyposis coli 2 FLJ22184 0.033 PREDICTED: hypothetical protein FLJ22184 LOC283999 0.032 hypothetical protein LOC283999 HABP4 0.032 hyaluronan binding protein 4 LOC100130897 0.030 PREDICTED: hypothetical protein LOC100130897 0.030 PREDICTED: hypothetical protein FLJ37078 0.030 hypothetical protein LOC222183 LOC100291533 0.029 PREDICTED: hypothetical protein XP_002347080 LOC100288002 0.029 PREDICTED: hypothetical protein XP_002342946 LOC100292057 0.029 PREDICTED: hypothetical protein XP_002345830 LOC100287121 0.029 PREDICTED: hypothetical protein LOC100287121 0.029 PREDICTED: hypothetical protein XP_002342511 SRRM1 0.029 serine/arginine repetitive matrix 1 LOC100291147 0.027 PREDICTED: hypothetical protein XP_002347883 LOC100289560 0.027 PREDICTED: hypothetical protein XP_002344188 LOC100127930 0.026 PREDICTED: hypothetical protein LOC100128511 0.026 PREDICTED: hypothetical protein LOC100132891 0.026 PREDICTED: hypothetical protein LOC100128511 0.026 PREDICTED: hypothetical protein LOC100132891 0.026 PREDICTED: hypothetical protein LOC100128511 0.026 PREDICTED: hypothetical protein LOC100132891 0.026 PREDICTED: hypothetical protein TTMA 0.026 hypothetical protein LOC645369Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
