
| Name: LOC100292347 | Sequence: fasta or formatted (104aa) | NCBI GI: 239755429 | |
|
Description: PREDICTED: hypothetical protein
| Not currently referenced in the text | ||
|
Composition:
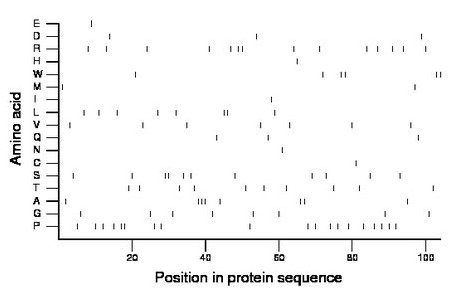
Amino acid Percentage Count Longest homopolymer A alanine 7.7 8 3 C cysteine 1.0 1 1 D aspartate 2.9 3 1 E glutamate 1.0 1 1 F phenylalanine 0.0 0 0 G glycine 7.7 8 1 H histidine 1.0 1 1 I isoleucine 1.0 1 1 K lysine 0.0 0 0 L leucine 7.7 8 2 M methionine 1.9 2 1 N asparagine 1.0 1 1 P proline 18.3 19 2 Q glutamine 2.9 3 1 R arginine 13.5 14 2 S serine 10.6 11 2 T threonine 9.6 10 1 V valine 6.7 7 1 W tryptophan 5.8 6 2 Y tyrosine 0.0 0 0 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein LOC100290593 1.000 PREDICTED: hypothetical protein XP_002347195 LOC100289632 1.000 PREDICTED: hypothetical protein XP_002343051 PSD 0.079 pleckstrin and Sec7 domain containing NACA 0.059 nascent polypeptide-associated complex alpha subuni... FAM189A1 0.050 hypothetical protein LOC23359 TCERG1 0.050 transcription elongation regulator 1 isoform 2 [Homo... TCERG1 0.050 transcription elongation regulator 1 isoform 1 [Homo... SOLH 0.050 small optic lobes LOC100131778 0.045 PREDICTED: hypothetical protein LOC100131646 0.045 PREDICTED: hypothetical protein LOC100131778 0.045 PREDICTED: hypothetical protein LOC100131778 0.045 PREDICTED: hypothetical protein SNAPC2 0.045 small nuclear RNA activating complex, polypeptide 2, ... SELV 0.040 selenoprotein V SRRM1 0.040 serine/arginine repetitive matrix 1 ANKRD34A 0.035 ankyrin repeat domain 34 IRAK1 0.035 interleukin-1 receptor-associated kinase 1 isoform 3... IRAK1 0.035 interleukin-1 receptor-associated kinase 1 isoform 2... IRAK1 0.035 interleukin-1 receptor-associated kinase 1 isoform 1... CDKN1C 0.035 cyclin-dependent kinase inhibitor 1C isoform b [Hom... CDKN1C 0.035 cyclin-dependent kinase inhibitor 1C isoform b [Hom... CDKN1C 0.035 cyclin-dependent kinase inhibitor 1C isoform a MUC2 0.035 mucin 2 precursor LOC100294420 0.035 PREDICTED: similar to mucin 2 DGKK 0.035 diacylglycerol kinase kappa IGSF9 0.035 immunoglobulin superfamily, member 9 isoform a [Hom... IGSF9 0.035 immunoglobulin superfamily, member 9 isoform b [Hom... SHANK1 0.035 SH3 and multiple ankyrin repeat domains 1 ANO8 0.030 anoctamin 8Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
