
| Name: LOC100293898 | Sequence: fasta or formatted (112aa) | NCBI GI: 239755362 | |
|
Description: PREDICTED: hypothetical protein
| Not currently referenced in the text | ||
|
Composition:
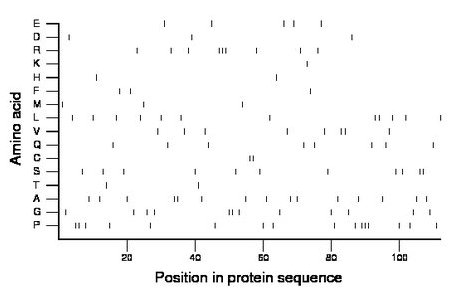
Amino acid Percentage Count Longest homopolymer A alanine 13.4 15 2 C cysteine 1.8 2 2 D aspartate 2.7 3 1 E glutamate 4.5 5 1 F phenylalanine 2.7 3 1 G glycine 10.7 12 2 H histidine 1.8 2 1 I isoleucine 0.0 0 0 K lysine 0.9 1 1 L leucine 10.7 12 2 M methionine 2.7 3 1 N asparagine 0.0 0 0 P proline 14.3 16 3 Q glutamine 7.1 8 1 R arginine 8.0 9 3 S serine 9.8 11 2 T threonine 1.8 2 1 V valine 7.1 8 2 W tryptophan 0.0 0 0 Y tyrosine 0.0 0 0 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein TBX1 0.243 T-box 1 isoform C LOC100293292 0.099 PREDICTED: hypothetical protein LOC100287612 0.099 PREDICTED: hypothetical protein XP_002342706 NRG1 0.054 neuregulin 1 isoform GGF2 EP300 0.045 E1A binding protein p300 LOC100128142 0.035 PREDICTED: hypothetical protein LOC100128142 0.035 PREDICTED: similar to RIKEN cDNA E130309D14 NR5A1 0.035 nuclear receptor subfamily 5, group A, member 1 [Hom... CEBPE 0.035 CCAAT/enhancer binding protein epsilon LOC100291863 0.030 PREDICTED: hypothetical protein XP_002345880 LOC100287186 0.030 PREDICTED: hypothetical protein LOC100130431 0.030 PREDICTED: hypothetical protein LOC100287186 0.030 PREDICTED: hypothetical protein XP_002342580 BAT3 0.025 HLA-B associated transcript-3 isoform b BAT3 0.025 HLA-B associated transcript-3 isoform b BAT3 0.025 HLA-B associated transcript-3 isoform b BAT3 0.025 HLA-B associated transcript-3 isoform a AMOT 0.025 angiomotin isoform 2 AMOT 0.025 angiomotin isoform 1 CBFA2T3 0.025 myeloid translocation gene on chromosome 16 isoform ... CBFA2T3 0.025 myeloid translocation gene on chromosome 16 isoform ... LOC100292447 0.025 PREDICTED: similar to coiled-coil domain containing... PPP1R12C 0.025 protein phosphatase 1, regulatory subunit 12C BCL9 0.025 B-cell CLL/lymphoma 9 CABIN1 0.020 calcineurin binding protein 1 KIAA0100 0.015 hypothetical protein LOC9703 NCOR2 0.015 nuclear receptor co-repressor 2 isoform 2 UNCX 0.015 UNC homeobox SIPA1 0.015 signal-induced proliferation-associated protein 1 [...Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
