
| Name: LOC100293175 | Sequence: fasta or formatted (115aa) | NCBI GI: 239755146 | |
|
Description: PREDICTED: hypothetical protein
| Not currently referenced in the text | ||
|
Composition:
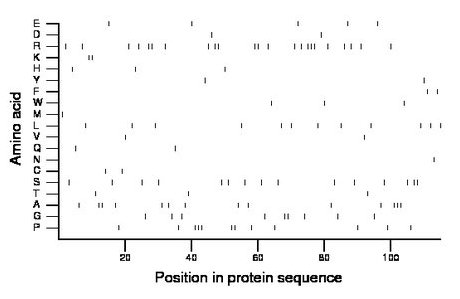
Amino acid Percentage Count Longest homopolymer A alanine 12.2 14 3 C cysteine 1.7 2 1 D aspartate 1.7 2 1 E glutamate 4.3 5 1 F phenylalanine 1.7 2 1 G glycine 7.8 9 2 H histidine 2.6 3 1 I isoleucine 0.0 0 0 K lysine 1.7 2 2 L leucine 10.4 12 1 M methionine 0.9 1 1 N asparagine 0.9 1 1 P proline 10.4 12 3 Q glutamine 1.7 2 1 R arginine 20.0 23 3 S serine 13.0 15 2 T threonine 2.6 3 1 V valine 1.7 2 1 W tryptophan 2.6 3 1 Y tyrosine 1.7 2 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein LOC100291458 0.991 PREDICTED: hypothetical protein XP_002347107 LOC100288099 0.991 PREDICTED: hypothetical protein XP_002342970 LOC100291932 0.061 PREDICTED: hypothetical protein XP_002345595 LOC100290844 0.061 PREDICTED: hypothetical protein XP_002348320 LOC100287739 0.061 PREDICTED: hypothetical protein XP_002342280 LOC100293956 0.047 PREDICTED: hypothetical protein LOC100289786 0.047 PREDICTED: hypothetical protein XP_002348005 LOC643355 0.047 PREDICTED: hypothetical protein LOC100287012 0.047 PREDICTED: hypothetical protein XP_002343725 LOC643355 0.047 PREDICTED: hypothetical protein LOC643355 0.042 PREDICTED: hypothetical protein LOC100290879 0.042 PREDICTED: hypothetical protein XP_002347499 LOC100286940 0.042 PREDICTED: hypothetical protein XP_002343317 FLJ37078 0.042 hypothetical protein LOC222183 SRRM2 0.037 splicing coactivator subunit SRm300 SART1 0.033 squamous cell carcinoma antigen recognized by T cell... CDC2L5 0.033 cell division cycle 2-like 5 isoform 1 CDC2L5 0.033 cell division cycle 2-like 5 isoform 2 LOC100170229 0.028 hypothetical protein LOC100170229 LOC100293818 0.028 PREDICTED: hypothetical protein LOC100134017 0.028 PREDICTED: hypothetical protein CEBPA 0.023 CCAAT/enhancer binding protein alpha NKAP 0.023 NFKB activating protein LOC100131673 0.023 PREDICTED: hypothetical protein WDR81 0.023 PREDICTED: WD repeat domain 81 LOC100131673 0.023 PREDICTED: hypothetical protein BPTF 0.023 bromodomain PHD finger transcription factor isoform ... BPTF 0.023 bromodomain PHD finger transcription factor isoform ... YTHDC1 0.023 splicing factor YT521-B isoform 2Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
