
| Name: LOC100290844 | Sequence: fasta or formatted (180aa) | NCBI GI: 239747803 | |
|
Description: PREDICTED: hypothetical protein XP_002348320
| Not currently referenced in the text | ||
|
Composition:
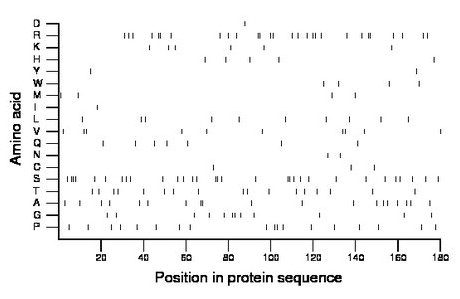
Amino acid Percentage Count Longest homopolymer A alanine 10.6 19 2 C cysteine 1.7 3 1 D aspartate 0.6 1 1 E glutamate 0.0 0 0 F phenylalanine 0.0 0 0 G glycine 6.7 12 2 H histidine 2.8 5 1 I isoleucine 0.6 1 1 K lysine 3.3 6 1 L leucine 5.6 10 1 M methionine 2.2 4 1 N asparagine 1.1 2 1 P proline 10.0 18 2 Q glutamine 3.9 7 1 R arginine 15.6 28 2 S serine 17.2 31 3 T threonine 9.4 17 1 V valine 5.6 10 2 W tryptophan 2.2 4 1 Y tyrosine 1.1 2 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein XP_002348320 LOC100291932 1.000 PREDICTED: hypothetical protein XP_002345595 LOC100287739 1.000 PREDICTED: hypothetical protein XP_002342280 SRRM2 0.065 splicing coactivator subunit SRm300 CCNL2 0.056 cyclin L2 isoform A LOC100133756 0.041 PREDICTED: hypothetical protein, partial CXorf67 0.041 hypothetical protein LOC340602 FUSIP1 0.041 FUS interacting protein (serine-arginine rich) 1 iso... LOC100293175 0.038 PREDICTED: hypothetical protein LOC100288382 0.038 PREDICTED: hypothetical protein XP_002342213 SFRS7 0.038 splicing factor, arginine/serine-rich 7 TRIOBP 0.038 TRIO and F-actin binding protein isoform 6 LOC100291458 0.035 PREDICTED: hypothetical protein XP_002347107 LOC100288099 0.035 PREDICTED: hypothetical protein XP_002342970 LOC100292133 0.032 PREDICTED: hypothetical protein LOC100294354 0.032 PREDICTED: hypothetical protein TRA2A 0.032 transformer-2 alpha ZRANB2 0.032 zinc finger protein 265 isoform 1 ADAMTS5 0.032 ADAM metallopeptidase with thrombospondin type 1 mo... SFRS6 0.029 arginine/serine-rich splicing factor 6 MICALL2 0.029 MICAL-like 2 isoform 1 PNRC1 0.029 proline-rich nuclear receptor coactivator 1 SR140 0.029 U2-associated SR140 protein ZRANB2 0.029 zinc finger protein 265 isoform 2 COL27A1 0.027 collagen, type XXVII, alpha 1 SFRS4 0.027 splicing factor, arginine/serine-rich 4 LOC100134017 0.027 PREDICTED: hypothetical protein MUC12 0.027 PREDICTED: mucin 12 LOC100291071 0.027 PREDICTED: hypothetical protein XP_002346951 LOC100287198 0.027 PREDICTED: hypothetical protein XP_002342815Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
