
| Name: LOC100293191 | Sequence: fasta or formatted (154aa) | NCBI GI: 239754761 | |
|
Description: PREDICTED: hypothetical protein
| Not currently referenced in the text | ||
|
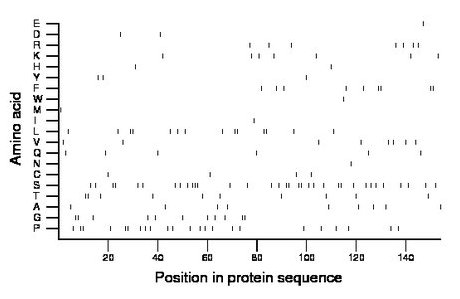
Composition:
Amino acid Percentage Count Longest homopolymer A alanine 5.8 9 1 C cysteine 2.6 4 1 D aspartate 1.3 2 1 E glutamate 0.6 1 1 F phenylalanine 5.8 9 2 G glycine 7.1 11 2 H histidine 1.3 2 1 I isoleucine 0.6 1 1 K lysine 4.5 7 1 L leucine 9.1 14 2 M methionine 0.6 1 1 N asparagine 0.6 1 1 P proline 14.9 23 2 Q glutamine 3.9 6 1 R arginine 4.5 7 1 S serine 22.1 34 3 T threonine 6.5 10 2 V valine 5.2 8 1 W tryptophan 0.6 1 1 Y tyrosine 1.9 3 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein LOC100290214 1.000 PREDICTED: hypothetical protein XP_002346983 LOC100288576 1.000 PREDICTED: hypothetical protein XP_002342845 LOC100288576 1.000 PREDICTED: hypothetical protein LOC100293166 0.403 PREDICTED: similar to ribosomal protein L36a LOC100290185 0.403 PREDICTED: hypothetical protein XP_002346982 LOC100288541 0.403 PREDICTED: hypothetical protein XP_002342844 LOC100288541 0.403 PREDICTED: similar to ribosomal protein L36a LOC100290274 0.334 PREDICTED: hypothetical protein XP_002346984 LOC100288645 0.334 PREDICTED: hypothetical protein XP_002342846 MAML2 0.055 mastermind-like 2 MAPK7 0.052 mitogen-activated protein kinase 7 isoform 1 MAPK7 0.052 mitogen-activated protein kinase 7 isoform 1 MAPK7 0.052 mitogen-activated protein kinase 7 isoform 1 MAPK7 0.052 mitogen-activated protein kinase 7 isoform 2 POLR2A 0.048 DNA-directed RNA polymerase II A GLTSCR1 0.048 glioma tumor suppressor candidate region gene 1 [Ho... SON 0.048 SON DNA-binding protein isoform F SON 0.048 SON DNA-binding protein isoform B SYNPO2L 0.048 synaptopodin 2-like isoform a SYNPO2L 0.048 synaptopodin 2-like isoform b TNKS 0.041 tankyrase, TRF1-interacting ankyrin-related ADP-ribo... PNRC1 0.041 proline-rich nuclear receptor coactivator 1 ABI1 0.041 abl-interactor 1 isoform a PRDM2 0.041 retinoblastoma protein-binding zinc finger protein i... PRDM2 0.041 retinoblastoma protein-binding zinc finger protein i... PRDM2 0.041 retinoblastoma protein-binding zinc finger protein i... DCX 0.041 doublecortin isoform c DCX 0.041 doublecortin isoform c DCX 0.041 doublecortin isoform aHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
