
| Name: LOC100292406 | Sequence: fasta or formatted (201aa) | NCBI GI: 239754544 | |
|
Description: PREDICTED: hypothetical protein
| Not currently referenced in the text | ||
|
Composition:
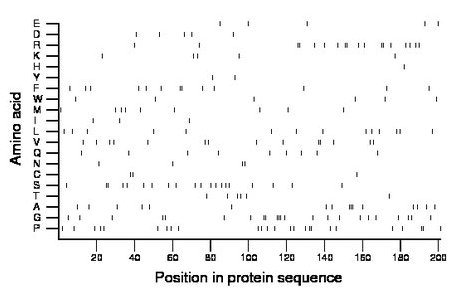
Amino acid Percentage Count Longest homopolymer A alanine 8.0 16 3 C cysteine 1.5 3 2 D aspartate 2.5 5 1 E glutamate 2.5 5 1 F phenylalanine 6.5 13 2 G glycine 11.4 23 3 H histidine 0.5 1 1 I isoleucine 1.5 3 1 K lysine 2.5 5 1 L leucine 7.0 14 1 M methionine 4.5 9 1 N asparagine 2.0 4 2 P proline 12.4 25 2 Q glutamine 4.5 9 1 R arginine 9.0 18 2 S serine 10.0 20 2 T threonine 3.0 6 1 V valine 7.0 14 2 W tryptophan 3.0 6 1 Y tyrosine 1.0 2 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein LOC100291258 1.000 PREDICTED: hypothetical protein XP_002346915 LOC100288357 1.000 PREDICTED: hypothetical protein XP_002342745 LOC100288357 1.000 PREDICTED: hypothetical protein LOC284988 0.035 PREDICTED: hypothetical protein ARID1B 0.030 AT rich interactive domain 1B (SWI1-like) isoform 2 ... ARID1B 0.030 AT rich interactive domain 1B (SWI1-like) isoform 1 ... ARID1B 0.030 AT rich interactive domain 1B (SWI1-like) isoform 3 ... LOC643355 0.023 PREDICTED: hypothetical protein LOC100288426 0.023 PREDICTED: hypothetical protein XP_002343430 LOC100291696 0.020 PREDICTED: hypothetical protein XP_002345913 LOC100291078 0.020 PREDICTED: hypothetical protein XP_002346792 LOC100293513 0.020 PREDICTED: hypothetical protein LOC100294343 0.020 PREDICTED: hypothetical protein XP_002344111 DIDO1 0.018 death inducer-obliterator 1 isoform c THSD1 0.018 thrombospondin type I domain-containing 1 isoform 2 ... THSD1 0.018 thrombospondin type I domain-containing 1 isoform 1 [... LZTS2 0.018 leucine zipper, putative tumor suppressor 2 DIAPH1 0.018 diaphanous 1 isoform 2 DIAPH1 0.018 diaphanous 1 isoform 1 LOC100289737 0.018 PREDICTED: hypothetical protein XP_002347183 LOC100288678 0.018 PREDICTED: hypothetical protein XP_002343039 FCHO1 0.015 FCH domain only 1 isoform c FCHO1 0.015 FCH domain only 1 isoform b FCHO1 0.015 FCH domain only 1 isoform b FCHO1 0.015 FCH domain only 1 isoform a LOC100291129 0.015 PREDICTED: hypothetical protein XP_002347815 SYN1 0.015 synapsin I isoform Ia SYN1 0.015 synapsin I isoform Ib LOC100293814 0.013 PREDICTED: hypothetical proteinHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
