
| Name: LOC100293457 | Sequence: fasta or formatted (134aa) | NCBI GI: 239754431 | |
|
Description: PREDICTED: hypothetical protein
| Not currently referenced in the text | ||
|
Composition:
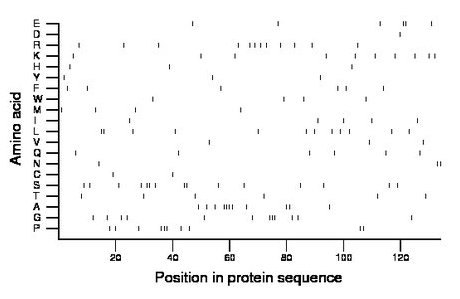
Amino acid Percentage Count Longest homopolymer A alanine 8.2 11 4 C cysteine 1.5 2 1 D aspartate 0.7 1 1 E glutamate 4.5 6 2 F phenylalanine 4.5 6 1 G glycine 9.0 12 3 H histidine 2.2 3 1 I isoleucine 3.7 5 1 K lysine 7.5 10 1 L leucine 9.0 12 2 M methionine 3.0 4 1 N asparagine 2.2 3 2 P proline 7.5 10 3 Q glutamine 4.5 6 1 R arginine 9.0 12 1 S serine 11.2 15 2 T threonine 4.5 6 1 V valine 2.2 3 1 W tryptophan 3.0 4 1 Y tyrosine 2.2 3 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein LOC100287407 0.764 PREDICTED: hypothetical protein XP_002342704 LOC100290769 0.654 PREDICTED: hypothetical protein XP_002346861 JUB 0.035 ajuba isoform 1 LOC651986 0.028 PREDICTED: similar to forkhead box L2 ARID1B 0.028 AT rich interactive domain 1B (SWI1-like) isoform 2 ... ARID1B 0.028 AT rich interactive domain 1B (SWI1-like) isoform 1 ... ARID1B 0.028 AT rich interactive domain 1B (SWI1-like) isoform 3 ... HCN4 0.024 hyperpolarization activated cyclic nucleotide-gated p... PHOX2B 0.020 paired-like homeobox 2b OSCAR 0.020 osteoclast-associated receptor isoform 1 LOC100288993 0.020 PREDICTED: hypothetical protein XP_002342967 GABBR1 0.016 gamma-aminobutyric acid (GABA) B receptor 1 isoform ... LOC388199 0.016 PREDICTED: hypothetical protein LOC388199 LOC388199 0.016 hypothetical protein LOC388199 LOC100127946 0.012 PREDICTED: hypothetical protein GUCY1A2 0.012 guanylate cyclase 1, soluble, alpha 2 UBQLN4 0.012 ataxin-1 ubiquitin-like interacting protein PLEKHG2 0.012 common-site lymphoma/leukemia guanine nucleotide ex... TNRC6B 0.012 trinucleotide repeat containing 6B isoform 2 TNRC6B 0.012 trinucleotide repeat containing 6B isoform 1 LOC100133915 0.012 PREDICTED: hypothetical protein HBS1L 0.008 Hsp70 subfamily B suppressor 1-like protein isoform... HBS1L 0.008 Hsp70 subfamily B suppressor 1-like protein isoform 1... C9orf66 0.008 hypothetical protein LOC157983 POU3F3 0.008 POU class 3 homeobox 3 AGAP2 0.008 centaurin, gamma 1 isoform PIKE-L GIYD1 0.008 GIY-YIG domain containing 1 isoform 2 GIYD1 0.008 GIY-YIG domain containing 1 isoform 1 LOC100294450 0.008 PREDICTED: hypothetical proteinHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
