
| Name: LOC100132738 | Sequence: fasta or formatted (1090aa) | NCBI GI: 239753106 | |
|
Description: PREDICTED: similar to hCG1817845
| Not currently referenced in the text | ||
Other entries for this name:
alt prot {572aa} PREDICTED: similar to hCG1817845 | |||
|
Composition:
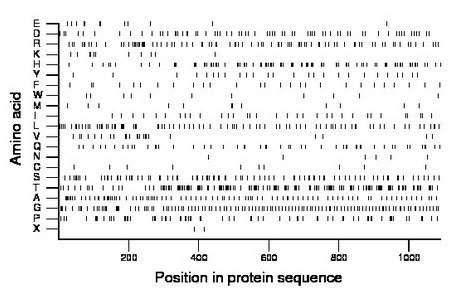
Amino acid Percentage Count Longest homopolymer A alanine 8.7 95 2 C cysteine 0.9 10 1 D aspartate 6.0 65 2 E glutamate 1.1 12 1 F phenylalanine 2.5 27 1 G glycine 10.8 118 2 H histidine 8.6 94 2 I isoleucine 3.0 33 1 K lysine 1.1 12 2 L leucine 7.8 85 3 M methionine 1.2 13 1 N asparagine 0.6 6 1 P proline 6.5 71 2 Q glutamine 4.8 52 1 R arginine 8.2 89 2 S serine 8.7 95 2 T threonine 13.7 149 2 V valine 2.3 25 2 W tryptophan 1.0 11 1 X unknown 0.2 2 1 Y tyrosine 2.4 26 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: similar to hCG1817845 LOC100132738 0.438 PREDICTED: similar to hCG1817845 FLG2 0.039 filaggrin family member 2 HRNR 0.022 hornerin FLG 0.019 filaggrin MUC12 0.013 PREDICTED: mucin 12 MUC12 0.011 PREDICTED: mucin 12, cell surface associated MUC6 0.011 mucin 6, gastric LOC100130716 0.009 PREDICTED: similar to mucin 11 MUC12 0.009 PREDICTED: mucin 12 MUC5AC 0.008 mucin 5AC LOC100133761 0.008 PREDICTED: similar to Mucin-6, partial LOC100133714 0.008 PREDICTED: similar to Mucin-12, partial FAM65A 0.008 hypothetical protein LOC79567 MUC5B 0.007 mucin 5, subtype B, tracheobronchial CATSPER1 0.005 sperm-associated cation channel 1 LOC729792 0.005 PREDICTED: hypothetical protein LOC729792 0.005 PREDICTED: hypothetical protein LOC729792 0.005 PREDICTED: hypothetical protein LOC100294501 0.005 PREDICTED: hypothetical protein LOC100294249 0.005 PREDICTED: hypothetical protein XP_002344067 LOC100294238 0.005 PREDICTED: hypothetical protein LOC100294204 0.005 PREDICTED: hypothetical protein LOC100294136 0.005 PREDICTED: hypothetical protein LOC100294091 0.005 PREDICTED: hypothetical protein LOC730401 0.005 PREDICTED: hypothetical protein DYRK1A 0.005 dual-specificity tyrosine-(Y)-phosphorylation regula... DYRK1A 0.005 dual-specificity tyrosine-(Y)-phosphorylation regula... PICALM 0.004 phosphatidylinositol-binding clathrin assembly prote... LOC100130716 0.004 PREDICTED: similar to Mucin-12Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
