
| Name: LOC100291218 | Sequence: fasta or formatted (203aa) | NCBI GI: 239750431 | |
|
Description: PREDICTED: hypothetical protein XP_002347361
| Not currently referenced in the text | ||
|
Composition:
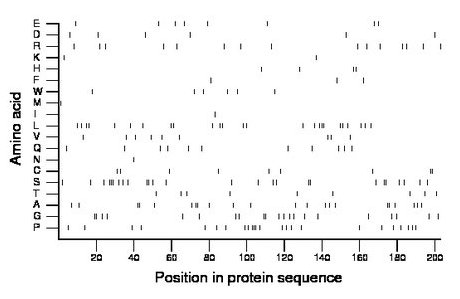
Amino acid Percentage Count Longest homopolymer A alanine 10.8 22 2 C cysteine 4.4 9 2 D aspartate 3.0 6 1 E glutamate 3.9 8 1 F phenylalanine 1.5 3 1 G glycine 10.3 21 2 H histidine 2.0 4 2 I isoleucine 0.5 1 1 K lysine 1.0 2 1 L leucine 12.3 25 3 M methionine 0.5 1 1 N asparagine 0.5 1 1 P proline 11.8 24 3 Q glutamine 5.4 11 1 R arginine 7.4 15 1 S serine 12.8 26 3 T threonine 4.4 9 1 V valine 4.4 9 1 W tryptophan 3.0 6 1 Y tyrosine 0.0 0 0 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein XP_002347361 TTC9B 0.030 tetratricopeptide repeat domain 9B HES3 0.030 hairy and enhancer of split 3 ADAMTS19 0.030 ADAM metallopeptidase with thrombospondin type 1 mo... LOC100292378 0.027 PREDICTED: hypothetical protein CRCT1 0.027 cysteine-rich C-terminal 1 KIF26A 0.027 kinesin family member 26A SHANK3 0.025 SH3 and multiple ankyrin repeat domains 3 WNK2 0.025 WNK lysine deficient protein kinase 2 RAPH1 0.020 Ras association and pleckstrin homology domains 1 is... CEBPA 0.020 CCAAT/enhancer binding protein alpha SPATA21 0.020 spermatogenesis associated 21 ZNF574 0.020 zinc finger protein 574 ROBO3 0.020 roundabout, axon guidance receptor, homolog 3 LOC100287250 0.017 PREDICTED: hypothetical protein XP_002344493 WASF2 0.017 WAS protein family, member 2 COLEC12 0.017 collectin sub-family member 12 FOXD1 0.017 forkhead box D1 CIC 0.017 capicua homolog OGFR 0.017 opioid growth factor receptor LOC100291699 0.017 PREDICTED: similar to hCG1991431 ZNF469 0.017 zinc finger protein 469 TESSP2 0.017 testis serine protease 2 ANKRD17 0.017 ankyrin repeat domain protein 17 isoform a ANKRD17 0.017 ankyrin repeat domain protein 17 isoform b LOC100292796 0.017 PREDICTED: hypothetical protein LOC100287763 0.017 PREDICTED: hypothetical protein LOC100287763 0.017 PREDICTED: hypothetical protein XP_002342429 CHST2 0.017 carbohydrate (N-acetylglucosamine-6-O) sulfotransfer... FBLIM1 0.017 filamin-binding LIM protein-1 isoform bHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
