
| Name: LOC100287643 | Sequence: fasta or formatted (103aa) | NCBI GI: 239749953 | |
|
Description: PREDICTED: hypothetical protein
| Not currently referenced in the text | ||
Other entries for this name:
alt mRNA {103aa} PREDICTED: hypothetical protein XP_002343106 | |||
|
Composition:
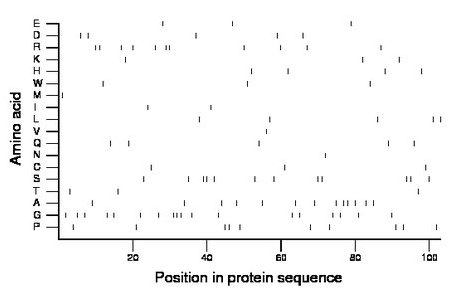
Amino acid Percentage Count Longest homopolymer A alanine 12.6 13 2 C cysteine 2.9 3 1 D aspartate 4.9 5 1 E glutamate 2.9 3 1 F phenylalanine 0.0 0 0 G glycine 17.5 18 3 H histidine 3.9 4 1 I isoleucine 1.9 2 1 K lysine 2.9 3 1 L leucine 4.9 5 1 M methionine 1.0 1 1 N asparagine 1.0 1 1 P proline 9.7 10 2 Q glutamine 4.9 5 1 R arginine 10.7 11 2 S serine 11.7 12 2 T threonine 2.9 3 1 V valine 1.0 1 1 W tryptophan 2.9 3 1 Y tyrosine 0.0 0 0 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein LOC100294012 1.000 PREDICTED: hypothetical protein LOC100287643 1.000 PREDICTED: hypothetical protein XP_002343106 LOC100289661 0.051 PREDICTED: hypothetical protein LOC100289661 0.051 PREDICTED: hypothetical protein XP_002343723 PP8961 0.045 hypothetical protein LOC650662 C20orf160 0.045 hypothetical protein LOC140706 WEE1 0.040 WEE1 tyrosine kinase isoform 1 LARP1 0.040 la related protein isoform 2 SPNS2 0.040 spinster homolog 2 C6orf174 0.035 hypothetical protein LOC387104 PRSS3 0.035 mesotrypsin isoform 1 preproprotein LOC100287063 0.035 PREDICTED: hypothetical protein XP_002342368 LOC100287063 0.030 PREDICTED: hypothetical protein ZNF469 0.030 zinc finger protein 469 ZDHHC8 0.030 zinc finger, DHHC domain containing 8 HRNR 0.025 hornerin LOC100291835 0.025 PREDICTED: hypothetical protein XP_002346266 LOC100293371 0.025 PREDICTED: hypothetical protein PHOX2A 0.025 paired-like homeobox 2a ADCY5 0.025 adenylate cyclase 5 GGN 0.025 gametogenetin MAGI2 0.025 membrane associated guanylate kinase, WW and PDZ dom... MLL4 0.025 myeloid/lymphoid or mixed-lineage leukemia 4 TTMA 0.025 hypothetical protein LOC645369 HNRNPU 0.020 heterogeneous nuclear ribonucleoprotein U isoform a ... ARID1B 0.020 AT rich interactive domain 1B (SWI1-like) isoform 2 ... ARID1B 0.020 AT rich interactive domain 1B (SWI1-like) isoform 1 ... ARID1B 0.020 AT rich interactive domain 1B (SWI1-like) isoform 3 ... RFTN1 0.020 raft-linking proteinHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
