
| Name: LOC100290144 | Sequence: fasta or formatted (111aa) | NCBI GI: 239749860 | |
|
Description: PREDICTED: hypothetical protein XP_002347189
| Not currently referenced in the text | ||
|
Composition:
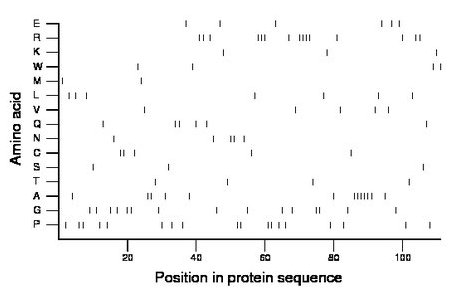
Amino acid Percentage Count Longest homopolymer A alanine 11.7 13 6 C cysteine 4.5 5 2 D aspartate 0.0 0 0 E glutamate 5.4 6 1 F phenylalanine 0.0 0 0 G glycine 13.5 15 2 H histidine 0.0 0 0 I isoleucine 0.0 0 0 K lysine 2.7 3 1 L leucine 6.3 7 1 M methionine 1.8 2 1 N asparagine 4.5 5 2 P proline 16.2 18 2 Q glutamine 5.4 6 2 R arginine 13.5 15 4 S serine 2.7 3 1 T threonine 3.6 4 1 V valine 4.5 5 1 W tryptophan 3.6 4 1 Y tyrosine 0.0 0 0 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein XP_002347189 LOC100291860 1.000 PREDICTED: hypothetical protein XP_002344548 ABL1 0.054 c-abl oncogene 1, receptor tyrosine kinase isoform a... ABL1 0.054 c-abl oncogene 1, receptor tyrosine kinase isoform b... PSMD3 0.050 proteasome 26S non-ATPase subunit 3 LOC729240 0.041 PREDICTED: FLJ40296 protein family member PRR20 0.041 proline rich 20 LOC729250 0.041 hypothetical protein LOC729250 LOC729246 0.041 hypothetical protein LOC729246 LOC729240 0.041 hypothetical protein LOC729240 LOC729233 0.041 hypothetical protein LOC729233 ZFP91 0.036 zinc finger protein 91 LOXL1 0.036 lysyl oxidase-like 1 preproprotein GLT8D4 0.036 glycosyltransferase 8 domain containing 4 SYN1 0.032 synapsin I isoform Ia SYN1 0.032 synapsin I isoform Ib C10orf114 0.032 hypothetical protein LOC399726 LOC100131591 0.032 PREDICTED: hypothetical protein LOC100291311 0.032 PREDICTED: hypothetical protein XP_002347439 LOC100289410 0.032 PREDICTED: hypothetical protein XP_002344274 STARD13 0.032 StAR-related lipid transfer (START) domain containin... STARD13 0.032 StAR-related lipid transfer (START) domain containin... STARD13 0.032 StAR-related lipid transfer (START) domain containin... PRB1 0.032 proline-rich protein BstNI subfamily 1 isoform 3 pre... LOC100130255 0.027 PREDICTED: hypothetical protein LOC100130255 0.027 PREDICTED: hypothetical protein LOC100130255 0.027 PREDICTED: hypothetical protein ALPK3 0.027 alpha-kinase 3 C9orf47 0.027 hypothetical protein LOC286223 isoform 1 BAHD1 0.027 bromo adjacent homology domain containing 1Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
