
| Name: LOC100290859 | Sequence: fasta or formatted (182aa) | NCBI GI: 239748061 | |
|
Description: PREDICTED: hypothetical protein XP_002346523
| Not currently referenced in the text | ||
|
Composition:
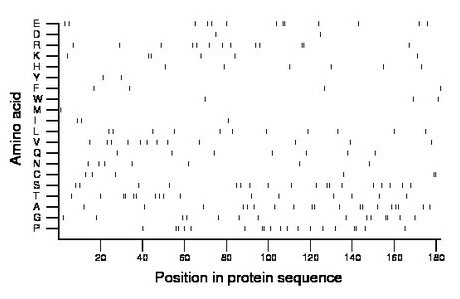
Amino acid Percentage Count Longest homopolymer A alanine 10.4 19 2 C cysteine 3.3 6 2 D aspartate 1.1 2 1 E glutamate 7.1 13 2 F phenylalanine 2.2 4 1 G glycine 7.7 14 2 H histidine 3.3 6 1 I isoleucine 1.6 3 1 K lysine 3.3 6 2 L leucine 6.0 11 1 M methionine 0.5 1 1 N asparagine 3.8 7 1 P proline 10.4 19 2 Q glutamine 3.8 7 1 R arginine 7.1 13 2 S serine 9.9 18 2 T threonine 8.8 16 2 V valine 6.6 12 1 W tryptophan 1.6 3 1 Y tyrosine 1.1 2 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein XP_002346523 LOC100288572 0.789 PREDICTED: hypothetical protein XP_002342361 LDB3 0.034 LIM domain binding 3 isoform 1 LOC338758 0.034 PREDICTED: hypothetical protein LOC338758 0.034 PREDICTED: hypothetical protein LOC338758 0.034 PREDICTED: hypothetical protein GPR152 0.031 G protein-coupled receptor 152 SEC16A 0.028 SEC16 homolog A TAF4 0.028 TBP-associated factor 4 EBF4 0.028 early B-cell factor 4 LIPE 0.028 hormone-sensitive lipase BSN 0.028 bassoon protein ANKRD56 0.028 ankyrin repeat domain 56 MGC87042 0.028 PREDICTED: similar to Six transmembrane epithelial ... MGC87042 0.028 PREDICTED: hypothetical protein LOC256227 EP400 0.028 E1A binding protein p400 SNX26 0.028 sorting nexin 26 LOC100290317 0.025 PREDICTED: hypothetical protein XP_002346944 PAK6 0.025 p21-activated kinase 6 PAK6 0.025 p21-activated kinase 6 PAK6 0.025 p21-activated kinase 6 MIA3 0.025 melanoma inhibitory activity family, member 3 [Homo... C15orf42 0.025 leucine-rich repeat kinase 1 ZNF831 0.025 zinc finger protein 831 WDR70 0.025 WD repeat domain 70 LOC727978 0.023 PREDICTED: hypothetical protein PCNT 0.023 pericentrin RERE 0.023 atrophin-1 like protein isoform b RERE 0.023 atrophin-1 like protein isoform a RERE 0.023 atrophin-1 like protein isoform aHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
