
| Name: LOC100294347 | Sequence: fasta or formatted (202aa) | NCBI GI: 239747249 | |
|
Description: PREDICTED: hypothetical protein
| Not currently referenced in the text | ||
|
Composition:
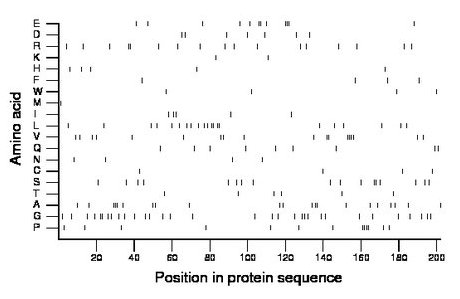
Amino acid Percentage Count Longest homopolymer A alanine 10.4 21 3 C cysteine 2.0 4 1 D aspartate 3.5 7 1 E glutamate 5.9 12 3 F phenylalanine 2.0 4 1 G glycine 15.8 32 2 H histidine 2.5 5 1 I isoleucine 2.5 5 2 K lysine 1.0 2 1 L leucine 10.4 21 2 M methionine 0.5 1 1 N asparagine 2.0 4 1 P proline 6.4 13 4 Q glutamine 4.5 9 1 R arginine 8.4 17 2 S serine 8.4 17 2 T threonine 3.0 6 1 V valine 8.9 18 4 W tryptophan 2.0 4 1 Y tyrosine 0.0 0 0 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein LOC100292079 0.992 PREDICTED: hypothetical protein XP_002345351 DDX51 0.026 DEAD (Asp-Glu-Ala-Asp) box polypeptide 51 LOC100292397 0.021 PREDICTED: hypothetical protein LOC100133142 0.021 PREDICTED: similar to zinc finger protein 208 [Homo... LOC100133142 0.021 PREDICTED: similar to zinc finger protein 208 [Homo... BSCL2 0.021 seipin isoform 1 BSCL2 0.021 seipin isoform 1 BSCL2 0.021 seipin isoform 2 THRAP3 0.018 thyroid hormone receptor associated protein 3 [Homo... LOC727838 0.018 PREDICTED: similar to cancer/testis antigen family ... ZNF784 0.018 zinc finger protein 784 FOXA1 0.013 forkhead box A1 CHD7 0.013 chromodomain helicase DNA binding protein 7 SYNGAP1 0.013 synaptic Ras GTPase activating protein 1 NFKBIZ 0.013 nuclear factor of kappa light polypeptide gene enhan... LOC100293327 0.013 PREDICTED: hypothetical protein LOC100291252 0.013 PREDICTED: hypothetical protein XP_002347634 LOC100287254 0.013 PREDICTED: hypothetical protein XP_002343484 ADCYAP1 0.010 adenylate cyclase activating polypeptide precursor ... ADCYAP1 0.010 adenylate cyclase activating polypeptide precursor ... LOC727838 0.010 PREDICTED: similar to cancer/testis antigen family ... NCAM1 0.010 neural cell adhesion molecule 1 isoform 3 RPGR 0.010 retinitis pigmentosa GTPase regulator isoform C [Hom... AATF 0.010 apoptosis antagonizing transcription factor CNPY4 0.010 canopy 4 homolog SKIV2L 0.010 superkiller viralicidic activity 2-like homolog [Hom... CDC42BPG 0.010 CDC42 binding protein kinase gamma (DMPK-like) [Hom... CACNA1I 0.010 calcium channel, voltage-dependent, T type, alpha 1I... CACNA1I 0.010 calcium channel, voltage-dependent, T type, alpha 1I...Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
