
| Name: C14orf181 | Sequence: fasta or formatted (166aa) | NCBI GI: 116642878 | |
|
Description: hypothetical protein LOC400223
| Not currently referenced in the text | ||
|
Composition:
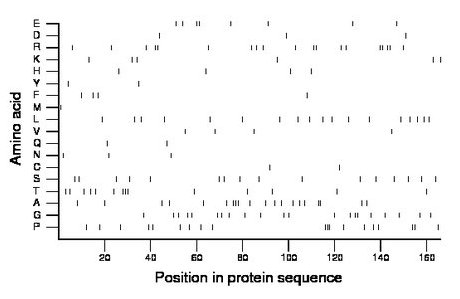
Amino acid Percentage Count Longest homopolymer A alanine 12.0 20 3 C cysteine 1.2 2 1 D aspartate 1.8 3 1 E glutamate 4.8 8 2 F phenylalanine 2.4 4 1 G glycine 12.7 21 2 H histidine 2.4 4 1 I isoleucine 0.0 0 0 K lysine 3.6 6 1 L leucine 10.8 18 1 M methionine 0.6 1 1 N asparagine 1.8 3 1 P proline 11.4 19 3 Q glutamine 1.2 2 1 R arginine 11.4 19 2 S serine 9.6 16 1 T threonine 8.4 14 3 V valine 2.4 4 1 W tryptophan 0.0 0 0 Y tyrosine 1.2 2 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 hypothetical protein LOC400223 CASKIN1 0.045 CASK interacting protein 1 LOC643355 0.038 PREDICTED: hypothetical protein NKX1-1 0.035 PREDICTED: NK1 homeobox 1 LOC100291627 0.032 PREDICTED: hypothetical protein XP_002345251 LOC100289867 0.032 PREDICTED: hypothetical protein XP_002348034 LOC100288231 0.032 PREDICTED: hypothetical protein XP_002343737 LOC643355 0.032 PREDICTED: hypothetical protein LOC643355 0.032 PREDICTED: hypothetical protein PLEKHG2 0.032 common-site lymphoma/leukemia guanine nucleotide ex... LOC100131009 0.029 PREDICTED: hypothetical protein LOC100131009 0.029 PREDICTED: hypothetical protein LOC100291624 0.029 PREDICTED: hypothetical protein XP_002345876 LOC100289712 0.029 PREDICTED: hypothetical protein XP_002346761 LOC100287455 0.029 PREDICTED: hypothetical protein XP_002342583 XIRP1 0.029 xin actin-binding repeat containing 1 CRAMP1L 0.026 Crm, cramped-like LOC100127930 0.026 PREDICTED: hypothetical protein LOC100292387 0.026 PREDICTED: similar to hemicentin 2 LOC100292865 0.026 PREDICTED: hypothetical protein HABP4 0.026 hyaluronan binding protein 4 LOC100293776 0.022 PREDICTED: hypothetical protein LOC100293575 0.022 PREDICTED: hypothetical protein LOC100290681 0.022 PREDICTED: hypothetical protein XP_002346859 LOC100131774 0.022 PREDICTED: hypothetical protein LOC100287330 0.022 PREDICTED: hypothetical protein XP_002342702 SNRPB 0.022 small nuclear ribonucleoprotein polypeptide B/B' iso... SNRPB 0.022 small nuclear ribonucleoprotein polypeptide B/B' isof... PHOX2A 0.022 paired-like homeobox 2a LTK 0.022 leukocyte receptor tyrosine kinase isoform 1 precurs...Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
