
| Name: LOC100289027 | Sequence: fasta or formatted (109aa) | NCBI GI: 239743776 | |
|
Description: PREDICTED: hypothetical protein XP_002344295
|
Referenced in:
| ||
|
Composition:
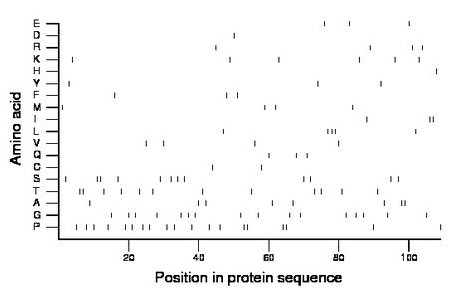
Amino acid Percentage Count Longest homopolymer A alanine 7.3 8 2 C cysteine 1.8 2 1 D aspartate 0.9 1 1 E glutamate 2.8 3 1 F phenylalanine 2.8 3 1 G glycine 14.7 16 1 H histidine 0.9 1 1 I isoleucine 2.8 3 2 K lysine 5.5 6 1 L leucine 4.6 5 3 M methionine 3.7 4 1 N asparagine 0.0 0 0 P proline 17.4 19 2 Q glutamine 2.8 3 1 R arginine 3.7 4 1 S serine 11.0 12 2 T threonine 11.0 12 2 V valine 3.7 4 1 W tryptophan 0.0 0 0 Y tyrosine 2.8 3 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein XP_002344295 CDK2AP2 0.782 cyclin-dependent kinase 2 associated protein 2 CDK2AP1 0.267 CDK2-associated protein 1 MAPK1IP1L 0.078 MAPK-interacting and spindle-stabilizing protein [Ho... DAAM1 0.073 dishevelled-associated activator of morphogenesis 1 ... MUC2 0.068 mucin 2 precursor LOC100294420 0.068 PREDICTED: similar to mucin 2 TMEM132E 0.058 transmembrane protein 132E RERE 0.053 atrophin-1 like protein isoform b RERE 0.053 atrophin-1 like protein isoform a RERE 0.053 atrophin-1 like protein isoform a FMN1 0.053 formin 1 COL17A1 0.053 alpha 1 type XVII collagen LOC100130360 0.049 PREDICTED: hypothetical protein MUC16 0.049 mucin 16 CCBE1 0.049 collagen and calcium binding EGF domains 1 NACA 0.049 nascent polypeptide-associated complex alpha subuni... LOC644794 0.049 PREDICTED: hypothetical protein LOC644794 0.049 PREDICTED: hypothetical protein LOC644794 0.049 PREDICTED: hypothetical protein LOC644794 0.049 PREDICTED: hypothetical protein NUP62 0.049 nucleoporin 62kDa NUP62 0.049 nucleoporin 62kDa NUP62 0.049 nucleoporin 62kDa NUP62 0.049 nucleoporin 62kDa ZMIZ1 0.049 retinoic acid induced 17 MED19 0.044 mediator complex subunit 19 ZNF828 0.044 zinc finger protein 828 COL1A1 0.044 alpha 1 type I collagen preproprotein GP1BA 0.044 platelet glycoprotein Ib alpha polypeptide precurso...Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
