
| Name: NAT14 | Sequence: fasta or formatted (206aa) | NCBI GI: 23943862 | |
|
Description: N-acetyltransferase 14
|
Referenced in: Additional Cell Cycle Functions
| ||
|
Composition:
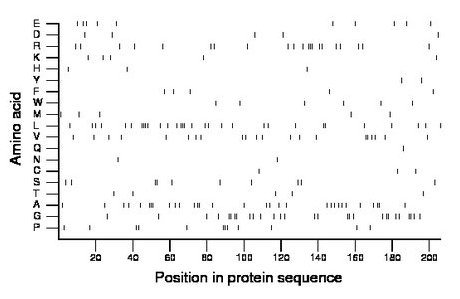
Amino acid Percentage Count Longest homopolymer A alanine 15.0 31 3 C cysteine 1.5 3 1 D aspartate 2.4 5 1 E glutamate 4.9 10 1 F phenylalanine 2.4 5 1 G glycine 13.6 28 2 H histidine 1.5 3 1 I isoleucine 0.0 0 0 K lysine 2.4 5 1 L leucine 15.0 31 4 M methionine 2.4 5 1 N asparagine 1.0 2 1 P proline 5.8 12 3 Q glutamine 0.5 1 1 R arginine 10.2 21 3 S serine 4.9 10 2 T threonine 2.4 5 1 V valine 10.2 21 2 W tryptophan 2.9 6 1 Y tyrosine 1.0 2 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 N-acetyltransferase 14 ELN 0.018 elastin isoform d precursor ELN 0.018 elastin isoform a precursor LOC100129169 0.015 PREDICTED: hypothetical protein LOC100129169 0.015 PREDICTED: hypothetical protein LOC100129169 0.015 PREDICTED: hypothetical protein PTPN23 0.015 protein tyrosine phosphatase, non-receptor type 23 [... GGT6 0.015 gamma-glutamyltransferase 6 isoform a GGT6 0.015 gamma-glutamyltransferase 6 isoform b KLHL34 0.015 kelch-like 34 ELN 0.015 elastin isoform e precursor ELN 0.015 elastin isoform c precursor ELN 0.015 elastin isoform b precursor LOC100291124 0.015 PREDICTED: hypothetical protein XP_002347542 MBD2 0.015 methyl-CpG binding domain protein 2 testis-specific i... MBD2 0.015 methyl-CpG binding domain protein 2 isoform 1 LOC100291147 0.013 PREDICTED: hypothetical protein XP_002347883 LOC100289560 0.013 PREDICTED: hypothetical protein XP_002344188 LOC100288473 0.013 PREDICTED: hypothetical protein XP_002343336 CLEC1B 0.013 C-type lectin domain family 1, member B isoform a [... CD72 0.013 CD72 molecule PLEKHH3 0.013 pleckstrin homology domain containing, family H (wit... LOC100294402 0.013 PREDICTED: similar to single Ig IL-1R-related molec... MSLN 0.013 mesothelin isoform 2 preproprotein MSLN 0.013 mesothelin isoform 1 preproprotein SEMA6B 0.013 semaphorin 6B precursor FCGRT 0.010 Fc fragment of IgG, receptor, transporter, alpha [H... FCGRT 0.010 Fc fragment of IgG, receptor, transporter, alpha [Hom... LOC100291835 0.010 PREDICTED: hypothetical protein XP_002346266 AGER 0.010 advanced glycosylation end product-specific receptor...Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
