
| Name: FANCE | Sequence: fasta or formatted (536aa) | NCBI GI: 11345454 | |
|
Description: Fanconi anemia, complementation group E
|
Referenced in: DNases, Recombination, and DNA Repair
| ||
|
Composition:
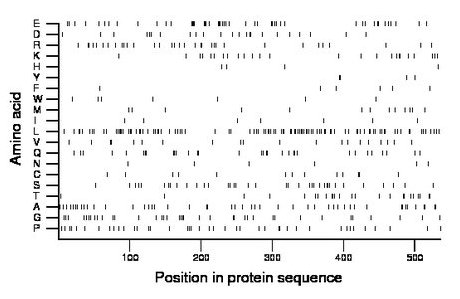
Amino acid Percentage Count Longest homopolymer A alanine 8.8 47 2 C cysteine 2.4 13 2 D aspartate 4.3 23 2 E glutamate 9.3 50 4 F phenylalanine 1.1 6 1 G glycine 7.1 38 2 H histidine 0.7 4 1 I isoleucine 1.3 7 1 K lysine 4.7 25 2 L leucine 19.2 103 3 M methionine 2.6 14 1 N asparagine 1.1 6 1 P proline 7.6 41 2 Q glutamine 6.0 32 2 R arginine 6.5 35 2 S serine 6.9 37 2 T threonine 4.5 24 2 V valine 3.7 20 2 W tryptophan 1.3 7 1 Y tyrosine 0.7 4 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 Fanconi anemia, complementation group E RGAG4 0.019 retrotransposon gag domain containing 4 ATRX 0.015 transcriptional regulator ATRX isoform 2 ATRX 0.015 transcriptional regulator ATRX isoform 1 KIF21B 0.014 kinesin family member 21B ANO8 0.014 anoctamin 8 ANP32E 0.014 acidic (leucine-rich) nuclear phosphoprotein 32 fami... ANP32E 0.014 acidic (leucine-rich) nuclear phosphoprotein 32 fam... ANP32E 0.014 acidic (leucine-rich) nuclear phosphoprotein 32 fam... VGF 0.014 VGF nerve growth factor inducible precursor NES 0.013 nestin PRG2 0.013 plasticity-related protein 2 OTUD3 0.013 OTU domain containing 3 LOC100170229 0.012 hypothetical protein LOC100170229 NEFH 0.012 neurofilament, heavy polypeptide 200kDa KIAA1211 0.012 hypothetical protein LOC57482 KIF21A 0.012 kinesin family member 21A BAZ1A 0.012 bromodomain adjacent to zinc finger domain, 1A isofo... BAZ1A 0.012 bromodomain adjacent to zinc finger domain, 1A isofo... BAT2D1 0.012 HBxAg transactivated protein 2 LOC100133599 0.011 PREDICTED: hypothetical protein GNAL 0.011 guanine nucleotide binding protein (G protein), alph... CCDC64B 0.011 coiled-coil domain containing 64B AHDC1 0.011 AT hook, DNA binding motif, containing 1 TSC2 0.011 tuberous sclerosis 2 isoform 4 TSC2 0.011 tuberous sclerosis 2 isoform 1 TSC2 0.011 tuberous sclerosis 2 isoform 5 CTDP1 0.011 CTD (carboxy-terminal domain, RNA polymerase II, pol... LOC100291206 0.011 PREDICTED: hypothetical protein XP_002347014 LOC100294088 0.011 PREDICTED: hypothetical protein, partialHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
