
| Name: LOC644634 | Sequence: fasta or formatted (169aa) | NCBI GI: 113411437 | |
|
Description: PREDICTED: similar to UPF0627 protein ENSP00000358171 isoform 1
| Not currently referenced in the text | ||
|
Composition:
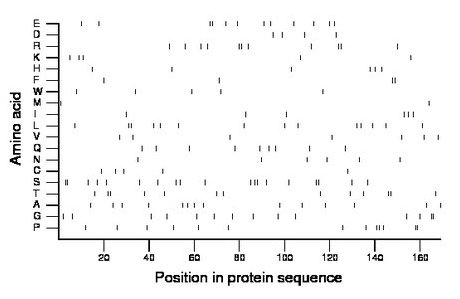
Amino acid Percentage Count Longest homopolymer A alanine 8.3 14 1 C cysteine 3.0 5 1 D aspartate 2.4 4 1 E glutamate 7.1 12 2 F phenylalanine 2.4 4 2 G glycine 8.3 14 2 H histidine 3.6 6 1 I isoleucine 3.6 6 1 K lysine 3.0 5 1 L leucine 8.3 14 2 M methionine 1.2 2 1 N asparagine 3.6 6 1 P proline 7.7 13 2 Q glutamine 5.3 9 1 R arginine 6.5 11 2 S serine 11.2 19 2 T threonine 7.7 13 2 V valine 4.1 7 1 W tryptophan 3.0 5 1 Y tyrosine 0.0 0 0 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: similar to UPF0627 protein ENSP000003581... LOC729587 0.860 PREDICTED: hypothetical protein LOC729574 0.860 PREDICTED: hypothetical protein LOC100133301 0.811 PREDICTED: hypothetical protein OSBPL10 0.024 oxysterol-binding protein-like protein 10 NID2 0.021 nidogen 2 precursor ZBTB20 0.018 zinc finger and BTB domain containing 20 ZFYVE26 0.015 zinc finger, FYVE domain containing 26 LOC729660 0.015 PREDICTED: similar to Putative uncharacterized prot... LOC731275 0.015 PREDICTED: similar to Putative uncharacterized prot... ZC3HAV1 0.012 zinc finger antiviral protein isoform 1 ZC3HAV1 0.012 zinc finger antiviral protein isoform 2 GCM1 0.012 glial cells missing homolog a GCM2 0.012 glial cells missing homolog 2 APLF 0.012 aprataxin and PNKP like factor PALM2-AKAP2 0.009 PALM2-AKAP2 protein isoform 2 PALM2-AKAP2 0.009 PALM2-AKAP2 protein isoform 1 LOC100293939 0.009 PREDICTED: hypothetical protein LOC100133167 0.009 PREDICTED: hypothetical protein LOC100291281 0.009 PREDICTED: hypothetical protein XP_002348078 LOC100289922 0.009 PREDICTED: hypothetical protein XP_002347871 LOC100288642 0.009 PREDICTED: hypothetical protein XP_002343774 ODZ4 0.009 odz, odd Oz/ten-m homolog 4 CANX 0.006 calnexin precursor CANX 0.006 calnexin precursor EXTL3 0.006 exostoses-like 3 TRAK1 0.006 OGT(O-Glc-NAc transferase)-interacting protein 106 ... TRAK1 0.006 OGT(O-Glc-NAc transferase)-interacting protein 106 K... ARHGAP21 0.006 Rho GTPase activating protein 21 FAM120C 0.006 hypothetical protein LOC54954Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
