
| Name: LOC100293939 | Sequence: fasta or formatted (145aa) | NCBI GI: 239757545 | |
|
Description: PREDICTED: hypothetical protein
| Not currently referenced in the text | ||
|
Composition:
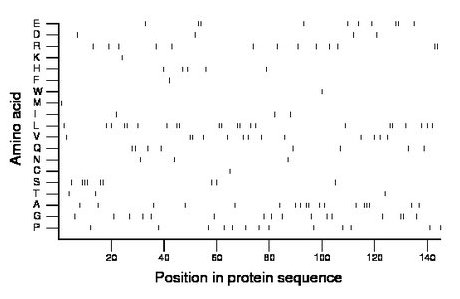
Amino acid Percentage Count Longest homopolymer A alanine 12.4 18 3 C cysteine 0.7 1 1 D aspartate 2.8 4 1 E glutamate 6.9 10 2 F phenylalanine 0.7 1 1 G glycine 11.0 16 2 H histidine 3.4 5 1 I isoleucine 2.1 3 1 K lysine 0.7 1 1 L leucine 15.2 22 2 M methionine 0.7 1 1 N asparagine 2.1 3 1 P proline 9.0 13 1 Q glutamine 5.5 8 2 R arginine 9.0 13 2 S serine 6.2 9 3 T threonine 2.1 3 1 V valine 9.0 13 2 W tryptophan 0.7 1 1 Y tyrosine 0.0 0 0 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein LOC100291281 1.000 PREDICTED: hypothetical protein XP_002348078 LOC100288642 1.000 PREDICTED: hypothetical protein XP_002343774 AUH 0.027 AU RNA binding protein/enoyl-Coenzyme A hydratase pre... LOC100129169 0.019 PREDICTED: hypothetical protein LOC100129169 0.019 PREDICTED: hypothetical protein LOC100129169 0.019 PREDICTED: hypothetical protein TNKS1BP1 0.019 tankyrase 1-binding protein 1 LOC100293087 0.015 PREDICTED: hypothetical protein LOC100127930 0.015 PREDICTED: hypothetical protein LOC729587 0.011 PREDICTED: hypothetical protein LOC729574 0.011 PREDICTED: hypothetical protein KCNH2 0.011 voltage-gated potassium channel, subfamily H, member... KCNH2 0.011 voltage-gated potassium channel, subfamily H, member ... ANKRD36 0.011 PREDICTED: ankyrin repeat domain 36 LOC644634 0.011 PREDICTED: similar to UPF0627 protein ENSP000003581... MMP11 0.011 matrix metalloproteinase 11 preproprotein SLC38A10 0.011 solute carrier family 38, member 10 isoform a RASAL1 0.011 RAS protein activator like 1 CP 0.008 ceruloplasmin precursor LOC100290189 0.008 PREDICTED: hypothetical protein XP_002347700 LOC100289086 0.008 PREDICTED: hypothetical protein XP_002344149 DVL3 0.008 dishevelled 3 LACTB 0.008 lactamase, beta isoform a LACTB 0.008 lactamase, beta isoform b SPOCD1 0.008 SPOC domain containing 1 LOC100127930 0.008 PREDICTED: hypothetical protein LOC100291385 0.008 PREDICTED: hypothetical protein XP_002347391 LOC100127930 0.008 PREDICTED: hypothetical protein LOC100287401 0.008 PREDICTED: hypothetical protein XP_002342373Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
