
| Name: LOC100131436 | Sequence: fasta or formatted (176aa) | NCBI GI: 169210715 | |
|
Description: PREDICTED: hypothetical protein
| Not currently referenced in the text | ||
Other entries for this name:
alt prot {176aa} PREDICTED: hypothetical protein | |||
|
Composition:
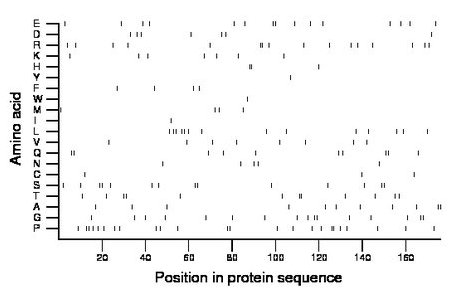
Amino acid Percentage Count Longest homopolymer A alanine 6.2 11 2 C cysteine 1.7 3 1 D aspartate 4.0 7 1 E glutamate 8.5 15 2 F phenylalanine 2.3 4 1 G glycine 9.1 16 2 H histidine 1.7 3 2 I isoleucine 0.6 1 1 K lysine 4.0 7 1 L leucine 8.0 14 2 M methionine 2.3 4 1 N asparagine 2.8 5 1 P proline 13.6 24 2 Q glutamine 5.7 10 2 R arginine 9.1 16 2 S serine 8.0 14 2 T threonine 6.8 12 2 V valine 4.5 8 1 W tryptophan 0.6 1 1 Y tyrosine 0.6 1 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein LOC100131436 0.991 PREDICTED: hypothetical protein AKT1S1 0.032 AKT1 substrate 1 (proline-rich) AKT1S1 0.032 AKT1 substrate 1 (proline-rich) AKT1S1 0.032 AKT1 substrate 1 (proline-rich) CTDP1 0.029 CTD (carboxy-terminal domain, RNA polymerase II, pol... CTDP1 0.029 CTD (carboxy-terminal domain, RNA polymerase II, pol... SH3PXD2B 0.023 SH3 and PX domains 2B LOC100131368 0.023 PREDICTED: similar to hCG1783157 LOC100291521 0.023 PREDICTED: hypothetical protein XP_002348052 LOC100289308 0.023 PREDICTED: hypothetical protein XP_002344215 HOXA4 0.020 homeobox A4 KIAA1217 0.020 sickle tail isoform 1 COL9A3 0.020 alpha 3 type IX collagen ZNF469 0.018 zinc finger protein 469 NCOA3 0.018 nuclear receptor coactivator 3 isoform a NCOA3 0.018 nuclear receptor coactivator 3 isoform b FLJ20021 0.018 PREDICTED: hypothetical protein FLJ20021 0.018 PREDICTED: hypothetical protein FLJ20021 0.018 PREDICTED: hypothetical protein ZMYND15 0.018 zinc finger, MYND-type containing 15 isoform 1 [Hom... ZMYND15 0.018 zinc finger, MYND-type containing 15 isoform 2 [Homo... C12orf68 0.018 hypothetical protein LOC387856 CDH4 0.015 cadherin 4, type 1 preproprotein LOC100289408 0.015 PREDICTED: hypothetical protein LOC100289408 0.015 PREDICTED: hypothetical protein XP_002344217 COQ10A 0.015 coenzyme Q10 homolog A isoform a PDCD7 0.015 programmed cell death 7 SS18 0.015 synovial sarcoma translocation, chromosome 18 isofor... NFATC4 0.015 nuclear factor of activated T-cells, cytoplasmic, c...Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
