
| Name: HINT2 | Sequence: fasta or formatted (163aa) | NCBI GI: 14211923 | |
|
Description: PKCI-1-related HIT protein
|
Referenced in:
| ||
|
Composition:
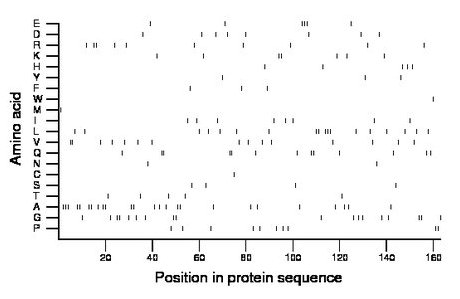
Amino acid Percentage Count Longest homopolymer A alanine 15.3 25 3 C cysteine 0.6 1 1 D aspartate 4.9 8 1 E glutamate 3.7 6 3 F phenylalanine 1.8 3 1 G glycine 11.0 18 2 H histidine 3.1 5 1 I isoleucine 4.9 8 1 K lysine 4.3 7 2 L leucine 11.0 18 3 M methionine 0.6 1 1 N asparagine 1.2 2 1 P proline 6.1 10 2 Q glutamine 8.0 13 2 R arginine 6.1 10 2 S serine 2.5 4 1 T threonine 3.1 5 1 V valine 9.2 15 2 W tryptophan 0.6 1 1 Y tyrosine 1.8 3 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PKCI-1-related HIT protein HINT1 0.466 histidine triad nucleotide binding protein 1 HINT3 0.074 histidine triad nucleotide binding protein 3 APTX 0.070 aprataxin isoform a APTX 0.070 aprataxin isoform c APTX 0.070 aprataxin isoform b APTX 0.057 aprataxin isoform d APTX 0.057 aprataxin isoform d FHIT 0.030 fragile histidine triad gene RBM28 0.017 RNA binding motif protein 28 TTN 0.017 titin isoform N2-A LOC100293374 0.017 PREDICTED: hypothetical protein LOC100287750 0.017 PREDICTED: hypothetical protein LOC100287750 0.017 PREDICTED: hypothetical protein XP_002342054 ANKFY1 0.017 ankyrin repeat and FYVE domain containing 1 isoform... LOC100133885 0.013 PREDICTED: hypothetical protein ZSWIM6 0.013 zinc finger, SWIM-type containing 6 NOL6 0.013 nucleolar protein family 6 gamma isoform NOL6 0.013 nucleolar protein family 6 alpha isoform KIAA0947 0.010 hypothetical protein LOC23379 LOC100292370 0.010 PREDICTED: hypothetical protein HNRNPM 0.010 heterogeneous nuclear ribonucleoprotein M isoform a ... HNRNPM 0.010 heterogeneous nuclear ribonucleoprotein M isoform b... EVPLL 0.007 envoplakin-like BMP6 0.007 bone morphogenetic protein 6 preproprotein CDC5L 0.007 CDC5-like SNX7 0.007 sorting nexin 7 isoform aHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
