
| Name: C9orf37 | Sequence: fasta or formatted (176aa) | NCBI GI: 63999355 | |
|
Description: chromosome 9 open reading frame 37
| Not currently referenced in the text | ||
|
Composition:
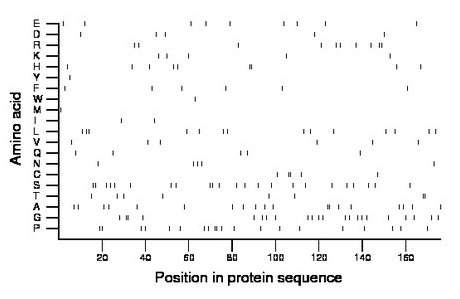
Amino acid Percentage Count Longest homopolymer A alanine 10.8 19 2 C cysteine 2.8 5 2 D aspartate 2.8 5 1 E glutamate 5.1 9 1 F phenylalanine 3.4 6 1 G glycine 12.5 22 2 H histidine 5.1 9 2 I isoleucine 1.1 2 1 K lysine 2.8 5 1 L leucine 8.0 14 2 M methionine 0.6 1 1 N asparagine 2.8 5 1 P proline 11.4 20 2 Q glutamine 2.8 5 1 R arginine 5.7 10 2 S serine 13.1 23 2 T threonine 4.5 8 2 V valine 3.4 6 1 W tryptophan 0.6 1 1 Y tyrosine 0.6 1 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 chromosome 9 open reading frame 37 C14orf178 0.104 hypothetical protein LOC283579 ZNF195 0.101 zinc finger protein 195 isoform 2 ZNF195 0.093 zinc finger protein 195 isoform 1 STAP2 0.087 signal transducing adaptor family member 2 isoform 1... C15orf40 0.070 hypothetical protein LOC123207 isoform d LOC100128260 0.070 PREDICTED: hCG_1742852 LOC100132815 0.070 PREDICTED: hypothetical protein LOC100128260 0.070 PREDICTED: hCG_1742852 LOC100132815 0.070 PREDICTED: hypothetical protein LOC100132815 0.070 PREDICTED: hypothetical protein CHEK2 0.067 protein kinase CHK2 isoform c LOC100288773 0.064 PREDICTED: hypothetical protein XP_002343911 LOC100128260 0.064 PREDICTED: hCG_1742852 UTY 0.064 tetratricopeptide repeat protein isoform 1 FLJ25363 0.058 hypothetical protein LOC401082 SULT1C2 0.055 sulfotransferase family, cytosolic, 1C, member 1 iso... GPR107 0.055 G protein-coupled receptor 107 isoform 1 SLC3A2 0.052 solute carrier family 3, member 2 isoform b SLC3A2 0.052 solute carrier family 3, member 2 isoform a SUGT1 0.052 suppressor of G2 allele of SKP1 isoform a USP19 0.052 ubiquitin thioesterase 19 GYG2 0.052 glycogenin 2 isoform b LOC100294407 0.049 PREDICTED: similar to hCG2039376 CNGA1 0.049 cyclic nucleotide gated channel alpha 1 isoform 1 [... LOC100128682 0.046 PREDICTED: hypothetical protein LOC100128682 LOC100128682 0.046 PREDICTED: hypothetical protein LOC100128682 LOC100128682 0.046 PREDICTED: hypothetical protein LOC100128682 PODXL 0.046 podocalyxin-like isoform 1 precursor REL 0.041 v-rel reticuloendotheliosis viral oncogene homolog [H...Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
