
| Name: EID2 | Sequence: fasta or formatted (236aa) | NCBI GI: 63175652 | |
|
Description: CREBBP/EP300 inhibitor 2
|
Referenced in:
| ||
|
Composition:
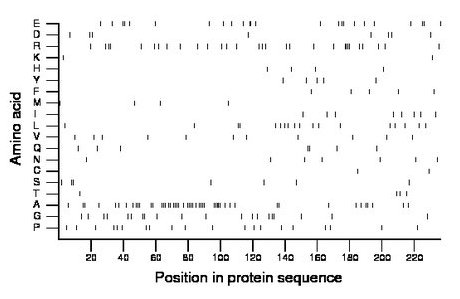
Amino acid Percentage Count Longest homopolymer A alanine 21.2 50 5 C cysteine 0.8 2 1 D aspartate 3.4 8 1 E glutamate 10.2 24 2 F phenylalanine 2.1 5 1 G glycine 8.5 20 2 H histidine 1.7 4 1 I isoleucine 3.4 8 1 K lysine 0.8 2 1 L leucine 7.6 18 2 M methionine 1.7 4 1 N asparagine 3.0 7 1 P proline 8.5 20 1 Q glutamine 3.0 7 2 R arginine 12.7 30 4 S serine 3.0 7 2 T threonine 1.7 4 1 V valine 4.7 11 1 W tryptophan 0.0 0 0 Y tyrosine 2.1 5 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 CREBBP/EP300 inhibitor 2 LOC100134071 0.232 PREDICTED: similar to EID2 protein LOC100290310 0.232 PREDICTED: hypothetical protein XP_002348293 LOC100287218 0.232 PREDICTED: hypothetical protein XP_002342306 EID2B 0.130 EP300 interacting inhibitor of differentiation 2B [H... EID1 0.070 CREBBP/EP300 inhibitor 1 SAMD1 0.057 sterile alpha motif domain containing 1 GNAS 0.057 GNAS complex locus XLas FAM48B1 0.048 hypothetical protein LOC100130302 HECA 0.039 headcase MARCKS 0.036 myristoylated alanine-rich protein kinase C substra... SOX21 0.036 SRY-box 21 C6orf174 0.036 hypothetical protein LOC387104 LOC728767 0.036 PREDICTED: hypothetical protein LOC728767 0.036 PREDICTED: hypothetical protein HOXA13 0.034 homeobox A13 SIRT1 0.034 sirtuin 1 isoform a BASP1 0.034 brain abundant, membrane attached signal protein 1 [... PAWR 0.034 PRKC, apoptosis, WT1, regulator LOC100290372 0.034 PREDICTED: hypothetical protein XP_002346788 LOC100288606 0.034 PREDICTED: hypothetical protein XP_002342652 MAGI2 0.032 membrane associated guanylate kinase, WW and PDZ dom... AMOT 0.032 angiomotin isoform 2 AMOT 0.032 angiomotin isoform 1 DACT3 0.032 thymus expressed gene 3-like TMPRSS13 0.032 transmembrane protease, serine 13 LOC100287688 0.032 PREDICTED: hypothetical protein XP_002342270 SOX3 0.032 SRY (sex determining region Y)-box 3 SPAG8 0.032 sperm associated antigen 8 isoform 1 FBRS 0.032 fibrosinHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
