
| Name: MRPL22 | Sequence: fasta or formatted (126aa) | NCBI GI: 62530388 | |
|
Description: mitochondrial ribosomal protein L22 isoform b
|
Referenced in:
| ||
Other entries for this name:
alt prot [206aa] mitochondrial ribosomal protein L22 isoform a | |||
|
Composition:
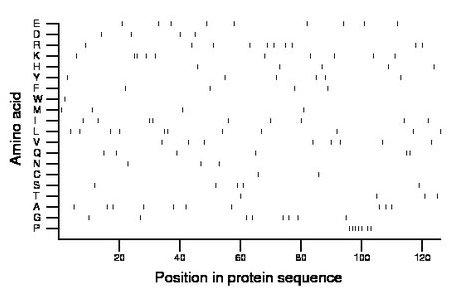
Amino acid Percentage Count Longest homopolymer A alanine 7.9 10 1 C cysteine 1.6 2 1 D aspartate 3.2 4 1 E glutamate 7.1 9 1 F phenylalanine 3.2 4 1 G glycine 6.3 8 1 H histidine 4.0 5 1 I isoleucine 7.1 9 2 K lysine 7.9 10 2 L leucine 8.7 11 2 M methionine 3.2 4 1 N asparagine 2.4 3 1 P proline 5.6 7 5 Q glutamine 4.8 6 2 R arginine 7.9 10 1 S serine 4.0 5 1 T threonine 3.2 4 1 V valine 6.3 8 1 W tryptophan 0.8 1 1 Y tyrosine 4.8 6 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 mitochondrial ribosomal protein L22 isoform b MRPL22 1.000 mitochondrial ribosomal protein L22 isoform a LOC100292865 0.026 PREDICTED: hypothetical protein CD96 0.017 CD96 antigen isoform 1 precursor CD96 0.017 CD96 antigen isoform 2 precursor PPM1B 0.017 protein phosphatase 1B isoform 3 PPM1B 0.017 protein phosphatase 1B isoform 1 ZC3H18 0.017 zinc finger CCCH-type containing 18 DKFZp686L13185 0.017 PREDICTED: hypothetical protein LOC401287 C11orf41 0.017 hypothetical protein LOC25758 PLA2G2E 0.017 phospholipase A2, group IIE PRR11 0.017 proline rich 11 CASKIN1 0.013 CASK interacting protein 1 TTN 0.013 titin isoform N2-A DKFZp686L13185 0.013 PREDICTED: hypothetical protein LOC401287 DKFZp686L13185 0.013 PREDICTED: hypothetical protein LOC401287 SOBP 0.013 sine oculis binding protein homolog ZFHX2 0.013 PREDICTED: zinc finger homeobox 2 ZFHX2 0.013 PREDICTED: zinc finger homeobox 2 ZFHX2 0.013 PREDICTED: zinc finger homeobox 2 LOC100129571 0.009 PREDICTED: similar to hCG1646049 LOC100129571 0.009 PREDICTED: similar to hCG1646049 LOC100129571 0.009 PREDICTED: similar to hCG1646049 PGK2 0.009 phosphoglycerate kinase 2 EFS 0.009 embryonal Fyn-associated substrate isoform 2 EFS 0.009 embryonal Fyn-associated substrate isoform 1 BCL9L 0.009 B-cell CLL/lymphoma 9-like MANEAL 0.009 mannosidase, endo-alpha-like isoform 3 MANEAL 0.009 mannosidase, endo-alpha-like isoform 1 RBL2 0.009 retinoblastoma-like 2 (p130)Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
