
| Name: SORCS1 | Sequence: fasta or formatted (1198aa) | NCBI GI: 61743975 | |
|
Description: SORCS receptor 1 isoform b
|
Referenced in:
| ||
Other entries for this name:
alt prot [1168aa] SORCS receptor 1 isoform a | |||
|
Composition:
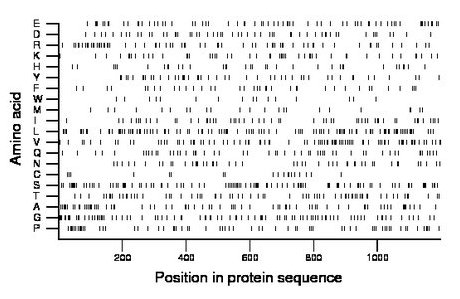
Amino acid Percentage Count Longest homopolymer A alanine 6.1 73 2 C cysteine 1.7 20 2 D aspartate 5.0 60 2 E glutamate 5.8 70 3 F phenylalanine 3.2 38 1 G glycine 7.6 91 3 H histidine 2.8 34 1 I isoleucine 4.8 58 2 K lysine 4.4 53 2 L leucine 9.3 112 2 M methionine 2.0 24 1 N asparagine 4.1 49 2 P proline 5.3 63 1 Q glutamine 4.5 54 2 R arginine 5.8 69 4 S serine 8.8 106 2 T threonine 5.8 70 2 V valine 7.4 89 2 W tryptophan 1.4 17 2 Y tyrosine 4.0 48 2 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 SORCS receptor 1 isoform b SORCS1 0.942 SORCS receptor 1 isoform a SORCS3 0.652 VPS10 domain receptor protein SORCS 3 SORCS2 0.410 VPS10 domain receptor protein SORCS 2 SORL1 0.095 sortilin-related receptor containing LDLR class A rep... SORT1 0.084 sortilin 1 preproprotein LOC100291634 0.007 PREDICTED: hypothetical protein XP_002346062 LOC100287232 0.007 PREDICTED: hypothetical protein XP_002342816 LOC100287232 0.007 PREDICTED: hypothetical protein LOC100129810 0.006 PREDICTED: hypothetical protein LOC100129810 0.006 PREDICTED: hypothetical protein LOC100291096 0.006 PREDICTED: hypothetical protein XP_002346952 LOC100129810 0.006 PREDICTED: hypothetical protein LOC100129810 0.006 PREDICTED: hypothetical protein JSRP1 0.005 junctional sarcoplasmic reticulum protein 1 LOC100292723 0.005 PREDICTED: hypothetical protein DTX1 0.005 deltex homolog 1 FOXH1 0.004 forkhead box H1 KAT5 0.004 K(lysine) acetyltransferase 5 isoform 3 RRP1 0.004 ribosomal RNA processing 1 homolog TNKS1BP1 0.004 tankyrase 1-binding protein 1 COL5A1 0.004 alpha 1 type V collagen preproprotein LMO2 0.004 LIM domain only 2 isoform 1 VASP 0.004 vasodilator-stimulated phosphoprotein LOC338758 0.004 PREDICTED: hypothetical protein LOC338758 0.004 PREDICTED: hypothetical protein LOC100130897 0.004 PREDICTED: hypothetical protein LOC338758 0.004 PREDICTED: hypothetical protein LOC100130897 0.004 PREDICTED: hypothetical protein MARCO 0.004 macrophage receptor with collagenous structureHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
