
| Name: C4orf46 | Sequence: fasta or formatted (113aa) | NCBI GI: 56606125 | |
|
Description: hypothetical protein LOC201725
| Not currently referenced in the text | ||
|
Composition:
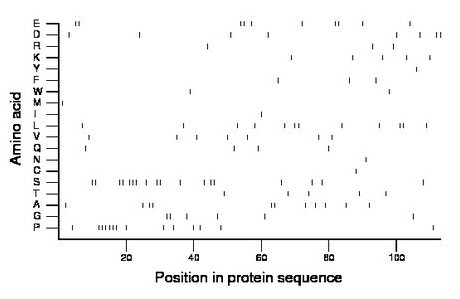
Amino acid Percentage Count Longest homopolymer A alanine 9.7 11 2 C cysteine 0.9 1 1 D aspartate 7.1 8 2 E glutamate 8.8 10 2 F phenylalanine 2.7 3 1 G glycine 5.3 6 2 H histidine 0.0 0 0 I isoleucine 0.9 1 1 K lysine 4.4 5 1 L leucine 10.6 12 2 M methionine 0.9 1 1 N asparagine 0.9 1 1 P proline 12.4 14 6 Q glutamine 3.5 4 1 R arginine 2.7 3 1 S serine 15.9 18 3 T threonine 4.4 5 1 V valine 6.2 7 1 W tryptophan 1.8 2 1 Y tyrosine 0.9 1 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 hypothetical protein LOC201725 LOC100129478 0.887 PREDICTED: hypothetical protein LOC100129478 0.887 PREDICTED: hypothetical protein LOC100129478 0.872 PREDICTED: hypothetical protein SYN2 0.064 synapsin II isoform IIa SYN2 0.064 synapsin II isoform IIb SMAD5 0.054 SMAD, mothers against DPP homolog 5 SMAD5 0.054 SMAD, mothers against DPP homolog 5 SMAD5 0.054 SMAD, mothers against DPP homolog 5 FUSSEL18 0.054 PREDICTED: functional smad suppressing element 18 [... FUSSEL18 0.054 PREDICTED: functional smad suppressing element 18 [... FUSSEL18 0.054 PREDICTED: functional smad suppressing element 18 [... RANBP9 0.049 RAN binding protein 9 LOC100293268 0.049 PREDICTED: hypothetical protein MYPOP 0.049 Myb protein P42POP KHDRBS2 0.049 KH domain-containing, RNA-binding, signal transduct... FOXG1 0.049 forkhead box G1 SFRS2IP 0.044 splicing factor, arginine/serine-rich 2, interactin... SPEM1 0.044 spermatid maturation 1 SMARCA5 0.039 SWI/SNF-related matrix-associated actin-dependent re... PPP3CB 0.039 protein phosphatase 3, catalytic subunit, beta isof... PPP3CB 0.039 protein phosphatase 3, catalytic subunit, beta isof... PPP3CB 0.039 protein phosphatase 3, catalytic subunit, beta isofo... BRD4 0.039 bromodomain-containing protein 4 isoform long ESPN 0.039 espin MEX3D 0.039 ring finger and KH domain containing 1 TMEM171 0.039 transmembrane protein 171 isoform 1 TMEM171 0.039 transmembrane protein 171 isoform 2 LDLRAD2 0.039 low density lipoprotein receptor class A domain con... TTN 0.039 titin isoform N2-AHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
