
| Name: UNC119 | Sequence: fasta or formatted (240aa) | NCBI GI: 4827048 | |
|
Description: unc119 (C.elegans) homolog isoform a
|
Referenced in: Non-Receptor Tyrosine Kinase Pathways
| ||
Other entries for this name:
alt prot [220aa] unc119 (C.elegans) homolog isoform b | |||
|
Composition:
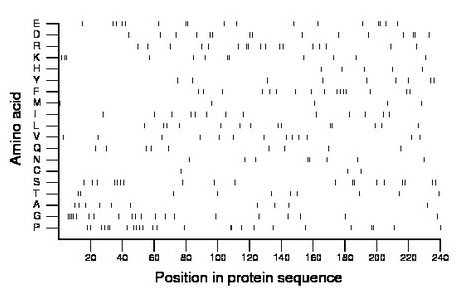
Amino acid Percentage Count Longest homopolymer A alanine 4.2 10 1 C cysteine 1.2 3 1 D aspartate 6.2 15 2 E glutamate 7.1 17 2 F phenylalanine 6.7 16 1 G glycine 8.3 20 4 H histidine 2.1 5 1 I isoleucine 5.4 13 1 K lysine 5.0 12 2 L leucine 5.8 14 2 M methionine 2.1 5 1 N asparagine 3.3 8 2 P proline 10.4 25 2 Q glutamine 3.8 9 1 R arginine 7.1 17 2 S serine 7.9 19 2 T threonine 4.2 10 1 V valine 5.4 13 1 W tryptophan 0.0 0 0 Y tyrosine 3.8 9 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 unc119 (C.elegans) homolog isoform a UNC119 0.838 unc119 (C.elegans) homolog isoform b UNC119B 0.523 unc-119 homolog B PDE6D 0.040 phosphodiesterase 6D COL1A1 0.032 alpha 1 type I collagen preproprotein BSN 0.030 bassoon protein PDCD7 0.025 programmed cell death 7 LOC100291634 0.025 PREDICTED: hypothetical protein XP_002346062 LOC100291096 0.025 PREDICTED: hypothetical protein XP_002346952 LOC100287232 0.025 PREDICTED: hypothetical protein XP_002342816 LOC100287232 0.025 PREDICTED: hypothetical protein DIAPH3 0.025 diaphanous homolog 3 isoform b DIAPH3 0.025 diaphanous homolog 3 isoform a COL6A1 0.025 collagen, type VI, alpha 1 precursor CCDC96 0.025 coiled-coil domain containing 96 HCN2 0.023 hyperpolarization activated cyclic nucleotide-gated... BAI1 0.023 brain-specific angiogenesis inhibitor 1 precursor [... COL16A1 0.023 alpha 1 type XVI collagen precursor EHBP1L1 0.023 tangerin FOXA3 0.023 forkhead box A3 LOC646509 0.021 PREDICTED: hypothetical protein FAM110C 0.021 hypothetical protein LOC642273 COL25A1 0.021 collagen, type XXV, alpha 1 isoform 1 COL25A1 0.021 collagen, type XXV, alpha 1 isoform 2 COL17A1 0.021 alpha 1 type XVII collagen SMARCA4 0.021 SWI/SNF-related matrix-associated actin-dependent r... SMARCA4 0.021 SWI/SNF-related matrix-associated actin-dependent r... SMARCA4 0.021 SWI/SNF-related matrix-associated actin-dependent r... SMARCA4 0.021 SWI/SNF-related matrix-associated actin-dependent r... SMARCA4 0.021 SWI/SNF-related matrix-associated actin-dependent r...Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
