
| Name: TMEM149 | Sequence: fasta or formatted (355aa) | NCBI GI: 13375913 | |
|
Description: transmembrane protein 149
| Not currently referenced in the text | ||
|
Composition:
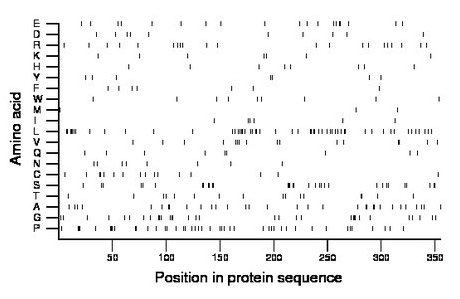
Amino acid Percentage Count Longest homopolymer A alanine 9.0 32 2 C cysteine 4.5 16 2 D aspartate 2.8 10 1 E glutamate 5.4 19 2 F phenylalanine 2.0 7 1 G glycine 9.0 32 3 H histidine 2.3 8 1 I isoleucine 1.7 6 1 K lysine 2.0 7 1 L leucine 14.9 53 4 M methionine 0.8 3 1 N asparagine 2.3 8 1 P proline 12.4 44 3 Q glutamine 2.5 9 2 R arginine 5.9 21 1 S serine 9.0 32 3 T threonine 4.5 16 2 V valine 4.5 16 2 W tryptophan 2.5 9 1 Y tyrosine 2.0 7 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 transmembrane protein 149 PEAR1 0.019 platelet endothelial aggregation receptor 1 KIAA0649 0.018 1A6/DRIM (down-regulated in metastasis) interacting ... LOC100293486 0.014 PREDICTED: similar to hCG2041452 LOC100291379 0.014 PREDICTED: hypothetical protein XP_002346556 LOC100288960 0.014 PREDICTED: hypothetical protein XP_002342392 NACA 0.014 nascent polypeptide-associated complex alpha subuni... TAF4 0.014 TBP-associated factor 4 PPP1R12B 0.014 protein phosphatase 1, regulatory (inhibitor) subuni... PPP1R12B 0.014 protein phosphatase 1, regulatory (inhibitor) subunit... SREBF1 0.014 sterol regulatory element binding transcription fact... SREBF1 0.014 sterol regulatory element binding transcription fact... TNK2 0.014 tyrosine kinase, non-receptor, 2 isoform 1 TNK2 0.014 tyrosine kinase, non-receptor, 2 isoform 2 RGS9 0.012 regulator of G-protein signaling 9 isoform 2 RGS9 0.012 regulator of G-protein signaling 9 isoform 1 TREM2 0.012 triggering receptor expressed on myeloid cells 2 prec... MGAM 0.012 maltase-glucoamylase CHKA 0.012 choline kinase alpha isoform b CHKA 0.012 choline kinase alpha isoform a BAT2 0.012 HLA-B associated transcript-2 KCP 0.012 cysteine rich BMP regulator 2 isoform 1 RIN3 0.012 Ras and Rab interactor 3 FMNL1 0.012 formin-like 1 LOC100289460 0.011 PREDICTED: hypothetical protein XP_002343919 LOC100291627 0.011 PREDICTED: hypothetical protein XP_002345251 LOC100289867 0.011 PREDICTED: hypothetical protein XP_002348034 LOC100288231 0.011 PREDICTED: hypothetical protein XP_002343737 GAK 0.011 cyclin G associated kinase ZNF828 0.011 zinc finger protein 828Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
