
| Name: ZNF428 | Sequence: fasta or formatted (188aa) | NCBI GI: 38788219 | |
|
Description: zinc finger protein 428
|
Referenced in: Kruppel-related Zinc Finger Proteins
| ||
|
Composition:
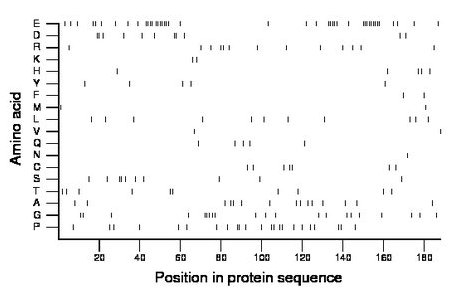
Amino acid Percentage Count Longest homopolymer A alanine 8.0 15 2 C cysteine 3.7 7 2 D aspartate 5.9 11 2 E glutamate 22.9 43 9 F phenylalanine 1.1 2 1 G glycine 11.2 21 3 H histidine 2.7 5 1 I isoleucine 0.0 0 0 K lysine 1.1 2 1 L leucine 5.9 11 1 M methionine 1.1 2 1 N asparagine 0.5 1 1 P proline 12.2 23 2 Q glutamine 2.7 5 1 R arginine 6.9 13 2 S serine 5.3 10 2 T threonine 5.3 10 2 V valine 1.1 2 1 W tryptophan 0.0 0 0 Y tyrosine 2.7 5 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 zinc finger protein 428 RTN4 0.071 reticulon 4 isoform B RTN4 0.071 reticulon 4 isoform D RTN4 0.071 reticulon 4 isoform A CCDC96 0.063 coiled-coil domain containing 96 ZBTB4 0.061 zinc finger and BTB domain containing 4 ZBTB4 0.061 zinc finger and BTB domain containing 4 PELP1 0.061 proline, glutamic acid and leucine rich protein 1 [... CHD3 0.058 chromodomain helicase DNA binding protein 3 isoform... RPGR 0.058 retinitis pigmentosa GTPase regulator isoform C [Hom... LIG1 0.056 DNA ligase I ARID4B 0.056 AT rich interactive domain 4B isoform 1 G3BP2 0.053 Ras-GTPase activating protein SH3 domain-binding pro... FUSSEL18 0.050 PREDICTED: functional smad suppressing element 18 [... FUSSEL18 0.050 PREDICTED: functional smad suppressing element 18 [... FUSSEL18 0.050 PREDICTED: functional smad suppressing element 18 [... TULP1 0.050 tubby like protein 1 CSRNP3 0.050 cysteine-serine-rich nuclear protein 3 MYT1 0.050 myelin transcription factor 1 HNRNPUL2 0.050 heterogeneous nuclear ribonucleoprotein U-like 2 [H... SCAF1 0.050 SR-related CTD-associated factor 1 CHIC1 0.050 cysteine-rich hydrophobic domain 1 ZNF629 0.050 zinc finger protein 629 ZSCAN10 0.050 zinc finger and SCAN domain containing 10 ZNF652 0.048 zinc finger protein 652 ZNF652 0.048 zinc finger protein 652 PPARGC1B 0.048 peroxisome proliferator-activated receptor gamma, co... LCP2 0.048 lymphocyte cytosolic protein 2 ARID3A 0.048 AT rich interactive domain 3A (BRIGHT- like) protein ... WNK2 0.045 WNK lysine deficient protein kinase 2Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
