
| Name: STRC | Sequence: fasta or formatted (1775aa) | NCBI GI: 24308527 | |
|
Description: stereocilin
|
Referenced in: Auditory and Vestibular Functions
| ||
|
Composition:
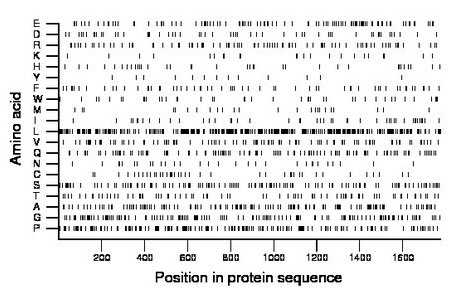
Amino acid Percentage Count Longest homopolymer A alanine 7.9 141 2 C cysteine 2.3 40 1 D aspartate 3.0 53 1 E glutamate 6.3 111 2 F phenylalanine 3.3 58 2 G glycine 8.1 143 2 H histidine 2.0 35 1 I isoleucine 2.6 47 1 K lysine 1.4 25 2 L leucine 18.3 325 11 M methionine 1.5 26 1 N asparagine 1.7 30 1 P proline 9.5 169 5 Q glutamine 6.5 116 2 R arginine 5.2 92 3 S serine 7.3 129 3 T threonine 4.7 83 2 V valine 5.5 98 2 W tryptophan 2.3 41 1 Y tyrosine 0.7 13 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 stereocilin OTOA 0.024 otoancorin isoform 3 OTOA 0.024 otoancorin isoform 2 OTOA 0.024 otoancorin isoform 1 PRR12 0.004 proline rich 12 TMUB1 0.004 transmembrane and ubiquitin-like domain containing 1... TMUB1 0.004 transmembrane and ubiquitin-like domain containing ... LOC100294292 0.004 PREDICTED: hypothetical protein XP_002344066 DAAM2 0.004 dishevelled associated activator of morphogenesis 2 ... FAM186A 0.003 family with sequence similarity 186, member A [Homo... LOC100293520 0.003 PREDICTED: hypothetical protein LOC100287207 0.003 PREDICTED: hypothetical protein XP_002342660 LOC100294237 0.003 PREDICTED: hypothetical protein LOC100294174 0.003 PREDICTED: hypothetical protein LOC100294124 0.003 PREDICTED: hypothetical protein LRP10 0.003 low density lipoprotein receptor-related protein 10 ... CCDC17 0.003 coiled-coil domain containing 17 FOXH1 0.003 forkhead box H1 COL4A5 0.003 type IV collagen alpha 5 isoform 2 precursor COL4A5 0.003 type IV collagen alpha 5 isoform 1 precursor COL4A5 0.003 type IV collagen alpha 5 isoform 3 precursor NRCAM 0.003 neuronal cell adhesion molecule isoform A precursor ... NRCAM 0.003 neuronal cell adhesion molecule isoform B precursor ... PGR 0.003 progesterone receptor LOC100293210 0.003 PREDICTED: hypothetical protein LOC100289267 0.003 PREDICTED: hypothetical protein XP_002342177 RTN2 0.003 reticulon 2 isoform A RTN2 0.003 reticulon 2 isoform B PRIC285 0.003 PPAR-alpha interacting complex protein 285 isoform ... PRIC285 0.003 PPAR-alpha interacting complex protein 285 isoform ...Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
