
| Name: LOC100134091 | Sequence: fasta or formatted (124aa) | NCBI GI: 239758069 | |
|
Description: PREDICTED: similar to FSHD region gene 1 isoform 2
| Not currently referenced in the text | ||
|
Composition:
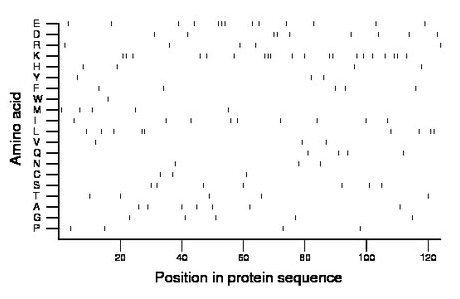
Amino acid Percentage Count Longest homopolymer A alanine 5.6 7 1 C cysteine 2.4 3 1 D aspartate 7.3 9 2 E glutamate 10.5 13 3 F phenylalanine 4.0 5 1 G glycine 4.0 5 1 H histidine 3.2 4 1 I isoleucine 7.3 9 1 K lysine 16.1 20 3 L leucine 7.3 9 2 M methionine 4.0 5 1 N asparagine 2.4 3 1 P proline 3.2 4 1 Q glutamine 3.2 4 1 R arginine 4.0 5 1 S serine 5.6 7 1 T threonine 4.0 5 1 V valine 2.4 3 1 W tryptophan 0.8 1 1 Y tyrosine 2.4 3 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: similar to FSHD region gene 1 isoform 2 ... FRG1 0.739 FSHD region gene 1 LOC100289524 0.704 PREDICTED: hypothetical protein XP_002343920 LOC100293552 0.683 PREDICTED: similar to hCG1992776 LOC100290870 0.683 PREDICTED: hypothetical protein XP_002346465 AKAP7 0.030 A-kinase anchor protein 7 isoform gamma SON 0.022 SON DNA-binding protein isoform F SON 0.022 SON DNA-binding protein isoform B RTF1 0.022 Paf1/RNA polymerase II complex component HMMR 0.017 hyaluronan-mediated motility receptor isoform d [Ho... HMMR 0.017 hyaluronan-mediated motility receptor isoform a [Ho... HMMR 0.017 hyaluronan-mediated motility receptor isoform c [Ho... HMMR 0.017 hyaluronan-mediated motility receptor isoform b [Ho... LOC100133599 0.013 PREDICTED: hypothetical protein ANKRD11 0.013 ankyrin repeat domain 11 UBE2Q1 0.013 ubiquitin-conjugating enzyme E2Q DST 0.013 dystonin isoform 1e precursor MAP1B 0.013 microtubule-associated protein 1B SMC1A 0.013 structural maintenance of chromosomes 1A KIF21A 0.013 kinesin family member 21A PALM2 0.013 paralemmin 2 isoform b PALM2 0.013 paralemmin 2 isoform a PALM2-AKAP2 0.013 PALM2-AKAP2 protein isoform 2 PALM2-AKAP2 0.013 PALM2-AKAP2 protein isoform 1 ASPH 0.009 aspartate beta-hydroxylase isoform d ASPH 0.009 aspartate beta-hydroxylase isoform e BAZ2A 0.009 bromodomain adjacent to zinc finger domain, 2A [Homo... TIMP2 0.009 TIMP metallopeptidase inhibitor 2 precursor ASPH 0.009 aspartate beta-hydroxylase isoform a RDX 0.009 radixinHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
