
| Name: LOC100293087 | Sequence: fasta or formatted (612aa) | NCBI GI: 239757595 | |
|
Description: PREDICTED: hypothetical protein
| Not currently referenced in the text | ||
|
Composition:
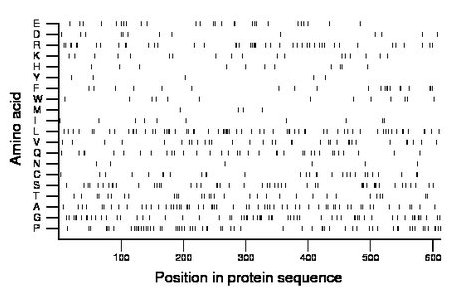
Amino acid Percentage Count Longest homopolymer A alanine 10.0 61 2 C cysteine 4.7 29 2 D aspartate 2.1 13 1 E glutamate 4.4 27 1 F phenylalanine 3.8 23 3 G glycine 10.3 63 2 H histidine 1.8 11 1 I isoleucine 1.6 10 1 K lysine 3.1 19 2 L leucine 10.8 66 3 M methionine 0.8 5 1 N asparagine 1.0 6 1 P proline 10.6 65 2 Q glutamine 5.7 35 2 R arginine 7.5 46 2 S serine 8.0 49 2 T threonine 5.2 32 2 V valine 5.6 34 3 W tryptophan 2.1 13 1 Y tyrosine 0.8 5 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein LOC100289735 0.375 PREDICTED: hypothetical protein XP_002348110 LOC100288372 0.375 PREDICTED: hypothetical protein XP_002343796 KRTAP4-9 0.013 PREDICTED: keratin associated protein 4-9 FLJ22184 0.012 PREDICTED: hypothetical protein FLJ22184 FLJ22184 0.012 PREDICTED: hypothetical protein LOC80164 FLJ22184 0.011 PREDICTED: hypothetical protein FLJ22184 XRCC1 0.010 X-ray repair cross complementing protein 1 KCNH4 0.010 potassium voltage-gated channel, subfamily H, member ... SRCAP 0.010 Snf2-related CBP activator protein INF2 0.010 inverted formin 2 isoform 2 INF2 0.010 inverted formin 2 isoform 1 C1orf70 0.010 hypothetical protein LOC339453 TP53 0.010 tumor protein p53 isoform b TP53 0.010 tumor protein p53 isoform c TP53 0.010 tumor protein p53 isoform a TP53 0.010 tumor protein p53 isoform a PELP1 0.010 proline, glutamic acid and leucine rich protein 1 [... WIPF3 0.009 WAS/WASL interacting protein family, member 3 [Homo... LOC100292845 0.008 PREDICTED: hypothetical protein LOC100289230 0.008 PREDICTED: hypothetical protein XP_002342600 WNK2 0.008 WNK lysine deficient protein kinase 2 FOXE3 0.008 forkhead box E3 LOC100134663 0.008 PREDICTED: hypothetical protein, partial RAPH1 0.008 Ras association and pleckstrin homology domains 1 is... LOC100134017 0.007 PREDICTED: hypothetical protein KDM4B 0.007 jumonji domain containing 2B RBM42 0.007 RNA binding motif protein 42 NPNT 0.007 nephronectin CPSF6 0.007 cleavage and polyadenylation specific factor 6, 68 ...Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
