
| Name: LOC100292950 | Sequence: fasta or formatted (82aa) | NCBI GI: 239757119 | |
|
Description: PREDICTED: hypothetical protein
| Not currently referenced in the text | ||
|
Composition:
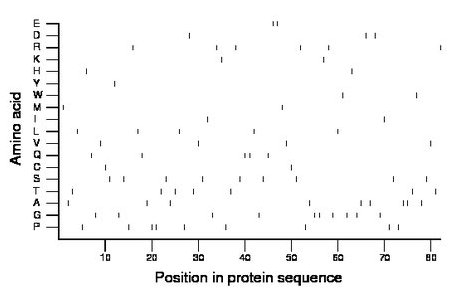
Amino acid Percentage Count Longest homopolymer A alanine 11.0 9 2 C cysteine 2.4 2 1 D aspartate 3.7 3 1 E glutamate 2.4 2 2 F phenylalanine 0.0 0 0 G glycine 12.2 10 2 H histidine 2.4 2 1 I isoleucine 2.4 2 1 K lysine 2.4 2 1 L leucine 6.1 5 1 M methionine 2.4 2 1 N asparagine 0.0 0 0 P proline 11.0 9 2 Q glutamine 6.1 5 2 R arginine 7.3 6 1 S serine 11.0 9 1 T threonine 8.5 7 1 V valine 4.9 4 1 W tryptophan 2.4 2 1 Y tyrosine 1.2 1 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein LOC100129571 0.034 PREDICTED: similar to hCG1646049 LOC100129571 0.034 PREDICTED: similar to hCG1646049 LOC100129571 0.034 PREDICTED: similar to hCG1646049 LOC100290372 0.020 PREDICTED: hypothetical protein XP_002346788 LOC100288606 0.020 PREDICTED: hypothetical protein XP_002342652 C7orf70 0.020 hypothetical protein LOC84792 C17orf65 0.020 hypothetical protein LOC339201 LOC100289661 0.013 PREDICTED: hypothetical protein LOC100289661 0.013 PREDICTED: hypothetical protein XP_002343723 FLJ10404 0.013 hypothetical protein LOC54540 TAX1BP1 0.013 Tax1 (human T-cell leukemia virus type I) binding pr... BRUNOL5 0.013 bruno-like 5, RNA binding protein isoform 2 MESP1 0.013 mesoderm posterior 1 KIF26A 0.013 kinesin family member 26A TRIM46 0.013 tripartite motif-containing 46 MUC16 0.013 mucin 16 LOC100132944 0.007 PREDICTED: similar to hCG2045457 LOC100292727 0.007 PREDICTED: hypothetical protein FLJ22184 0.007 PREDICTED: hypothetical protein FLJ22184 LOC647166 0.007 PREDICTED: similar to hCG28707 ZNF831 0.007 zinc finger protein 831 GAB1 0.007 GRB2-associated binding protein 1 isoform b GAB1 0.007 GRB2-associated binding protein 1 isoform a LOC100131436 0.007 PREDICTED: hypothetical protein ATP5B 0.007 mitochondrial ATP synthase beta subunit precursor [H...Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
