
| Name: LOC100294047 | Sequence: fasta or formatted (147aa) | NCBI GI: 239757007 | |
|
Description: PREDICTED: hypothetical protein
| Not currently referenced in the text | ||
|
Composition:
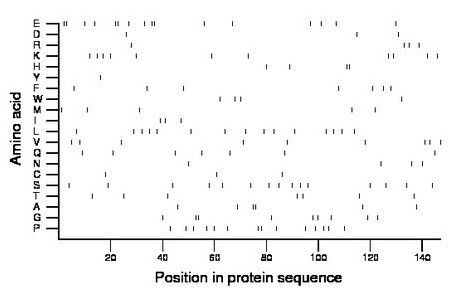
Amino acid Percentage Count Longest homopolymer A alanine 4.1 6 2 C cysteine 2.0 3 1 D aspartate 2.0 3 1 E glutamate 10.9 16 2 F phenylalanine 4.8 7 1 G glycine 6.1 9 2 H histidine 2.7 4 2 I isoleucine 2.0 3 1 K lysine 7.5 11 1 L leucine 10.2 15 1 M methionine 3.4 5 1 N asparagine 2.7 4 1 P proline 9.5 14 2 Q glutamine 4.8 7 1 R arginine 2.7 4 1 S serine 10.2 15 1 T threonine 4.8 7 1 V valine 6.1 9 1 W tryptophan 2.7 4 1 Y tyrosine 0.7 1 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein LOC100291094 1.000 PREDICTED: hypothetical protein XP_002347881 LOC100289436 1.000 PREDICTED: hypothetical protein XP_002344185 CHD8 0.018 chromodomain helicase DNA binding protein 8 SIK1 0.014 SNF1-like kinase RSBN1L 0.014 round spermatid basic protein 1-like C2orf71 0.011 hypothetical protein LOC388939 CAND2 0.011 TBP-interacting protein isoform 1 CAND2 0.011 TBP-interacting protein isoform 2 TAOK2 0.011 TAO kinase 2 isoform 2 ATF6B 0.011 activating transcription factor 6 beta isoform b [H... ATF6B 0.011 activating transcription factor 6 beta isoform a [Ho... C16orf5 0.011 cell death inducing protein ADAM10 0.011 ADAM metallopeptidase domain 10 LOC100128064 0.011 PREDICTED: hypothetical protein LOC100128064 0.011 PREDICTED: hypothetical protein PNPLA1 0.011 patatin-like phospholipase domain containing 1 isof... PNPLA1 0.011 patatin-like phospholipase domain containing 1 isof... PNPLA1 0.011 patatin-like phospholipase domain containing 1 isof... LOC100128064 0.007 PREDICTED: hypothetical protein ZNF469 0.007 zinc finger protein 469 ATXN2 0.007 ataxin 2 COQ3 0.007 hexaprenyldihydroxybenzoate methyltransferase [Homo... ABCC10 0.007 ATP-binding cassette, sub-family C, member 10 VPS13B 0.007 vacuolar protein sorting 13B isoform 5 VPS13B 0.007 vacuolar protein sorting 13B isoform 1 LOC644844 0.007 hypothetical protein LOC644844 VAX2 0.007 ventral anterior homeobox 2 KIF18B 0.007 kinesin family member 18B STX18 0.007 syntaxin 18Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
