
| Name: LOC100292353 | Sequence: fasta or formatted (247aa) | NCBI GI: 239756683 | |
|
Description: PREDICTED: hypothetical protein
| Not currently referenced in the text | ||
|
Composition:
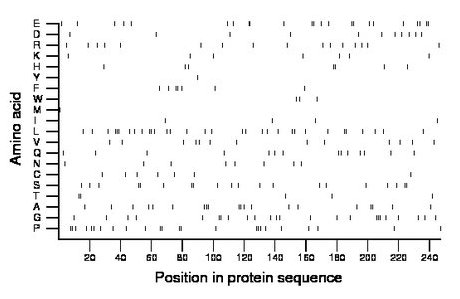
Amino acid Percentage Count Longest homopolymer A alanine 8.1 20 2 C cysteine 2.8 7 1 D aspartate 4.5 11 1 E glutamate 9.3 23 2 F phenylalanine 2.8 7 2 G glycine 9.7 24 3 H histidine 2.8 7 2 I isoleucine 1.6 4 1 K lysine 2.8 7 1 L leucine 13.0 32 3 M methionine 0.4 1 1 N asparagine 3.2 8 1 P proline 9.7 24 2 Q glutamine 5.7 14 1 R arginine 6.5 16 1 S serine 8.1 20 2 T threonine 2.0 5 2 V valine 5.3 13 1 W tryptophan 1.2 3 1 Y tyrosine 0.4 1 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein LOC100290241 0.548 PREDICTED: hypothetical protein XP_002347740 LOC100288617 0.414 PREDICTED: hypothetical protein XP_002343538 RP1L1 0.014 retinitis pigmentosa 1-like 1 TTLL7 0.014 tubulin tyrosine ligase-like family, member 7 KIAA1211 0.012 hypothetical protein LOC57482 ZNF497 0.012 zinc finger protein 497 FOXL1 0.010 forkhead box L1 SNRNP70 0.010 U1 small nuclear ribonucleoprotein 70 kDa RPAP1 0.010 RNA polymerase II associated protein 1 EPDR1 0.008 ependymin related protein 1 precursor LOC100293662 0.008 PREDICTED: hypothetical protein LOC100289444 0.008 PREDICTED: hypothetical protein LOC100289444 0.008 PREDICTED: hypothetical protein XP_002342754 LOC100289444 0.008 PREDICTED: hypothetical protein BNIP1 0.008 BCL2/adenovirus E1B 19kD interacting protein 1 isof... CASC5 0.006 cancer susceptibility candidate 5 isoform 2 CASC5 0.006 cancer susceptibility candidate 5 isoform 1 OTOG 0.006 PREDICTED: otogelin isoform 1 OTOG 0.006 PREDICTED: otogelin isoform 2 OTOG 0.006 PREDICTED: otogelin isoform 2 OTOG 0.006 PREDICTED: otogelin isoform 1 OTOG 0.006 PREDICTED: otogelin isoform 2 OTOG 0.006 PREDICTED: otogelin isoform 1 SLK 0.006 serine/threonine kinase 2 WIPF3 0.006 WAS/WASL interacting protein family, member 3 [Homo... CCDC64 0.006 coiled-coil domain containing 64 BNIP1 0.006 BCL2/adenovirus E1B 19kD interacting protein 1 isof... COL9A3 0.006 alpha 3 type IX collagen LOC100294103 0.006 PREDICTED: hypothetical proteinHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
