
| Name: LOC100291632 | Sequence: fasta or formatted (152aa) | NCBI GI: 239755323 | |
|
Description: PREDICTED: hypothetical protein XP_002344527
| Not currently referenced in the text | ||
|
Composition:
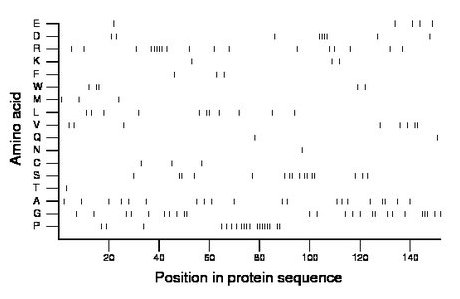
Amino acid Percentage Count Longest homopolymer A alanine 13.2 20 2 C cysteine 2.0 3 1 D aspartate 5.9 9 4 E glutamate 3.3 5 1 F phenylalanine 2.0 3 1 G glycine 16.4 25 3 H histidine 0.0 0 0 I isoleucine 0.0 0 0 K lysine 2.0 3 1 L leucine 7.2 11 2 M methionine 2.0 3 1 N asparagine 0.7 1 1 P proline 12.5 19 6 Q glutamine 1.3 2 1 R arginine 11.8 18 5 S serine 10.5 16 2 T threonine 0.7 1 1 V valine 5.3 8 2 W tryptophan 3.3 5 2 Y tyrosine 0.0 0 0 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein XP_002344527 LOC100288471 1.000 PREDICTED: hypothetical protein LOC100288471 1.000 PREDICTED: hypothetical protein XP_002343034 ZIC5 0.088 zinc finger protein of the cerebellum 5 WIPF3 0.071 WAS/WASL interacting protein family, member 3 [Homo... FNBP4 0.071 formin binding protein 4 PCLO 0.071 piccolo isoform 2 PCLO 0.071 piccolo isoform 1 FOXK1 0.068 forkhead box K1 KDM6B 0.068 lysine (K)-specific demethylase 6B RANBP9 0.064 RAN binding protein 9 MMP21 0.064 matrix metalloproteinase 21 preproprotein PDE4D 0.061 phosphodiesterase 4D isoform 1 LOC100131102 0.061 PREDICTED: hypothetical protein RAPH1 0.061 Ras association and pleckstrin homology domains 1 is... LOC100131102 0.061 PREDICTED: hypothetical protein FMNL2 0.061 formin-like 2 WHAMM 0.061 WAS protein homolog associated with actin, golgi me... ZFHX4 0.061 zinc finger homeodomain 4 ACR 0.061 acrosin precursor WIPF1 0.057 WAS/WASL interacting protein family, member 1 [Homo... WIPF1 0.057 WAS/WASL interacting protein family, member 1 TRRAP 0.057 transformation/transcription domain-associated protei... FMN1 0.057 formin 1 FUSSEL18 0.057 PREDICTED: functional smad suppressing element 18 [... FLJ20021 0.057 PREDICTED: hypothetical protein LOC645321 0.057 PREDICTED: hypothetical protein FUSSEL18 0.057 PREDICTED: functional smad suppressing element 18 [... FLJ20021 0.057 PREDICTED: hypothetical protein LOC645321 0.057 PREDICTED: hypothetical proteinHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
