
| Name: LOC647163 | Sequence: fasta or formatted (135aa) | NCBI GI: 239754316 | |
|
Description: PREDICTED: hypothetical protein
| Not currently referenced in the text | ||
Other entries for this name:
alt mRNA {135aa} PREDICTED: hypothetical protein alt mRNA {135aa} PREDICTED: hypothetical protein | |||
|
Composition:
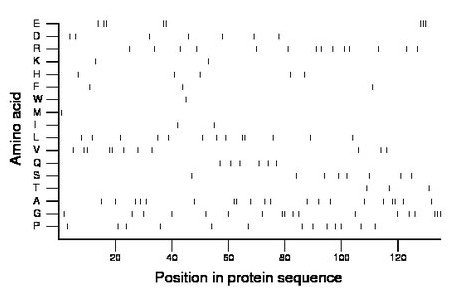
Amino acid Percentage Count Longest homopolymer A alanine 14.8 20 2 C cysteine 0.0 0 0 D aspartate 5.2 7 1 E glutamate 5.9 8 3 F phenylalanine 2.2 3 1 G glycine 13.3 18 3 H histidine 3.7 5 1 I isoleucine 1.5 2 1 K lysine 1.5 2 1 L leucine 9.6 13 2 M methionine 0.7 1 1 N asparagine 0.0 0 0 P proline 9.6 13 1 Q glutamine 4.4 6 1 R arginine 10.4 14 1 S serine 5.9 8 1 T threonine 2.2 3 1 V valine 8.1 11 2 W tryptophan 0.7 1 1 Y tyrosine 0.0 0 0 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein LOC647163 1.000 PREDICTED: hypothetical protein LOC647163 1.000 PREDICTED: hypothetical protein LOC100293502 0.065 PREDICTED: hypothetical protein LOC100289250 0.065 PREDICTED: hypothetical protein XP_002342089 LOC100293818 0.057 PREDICTED: hypothetical protein LOC100290009 0.049 PREDICTED: hypothetical protein XP_002346608 LOC100288104 0.049 PREDICTED: hypothetical protein XP_002342450 SYN1 0.036 synapsin I isoform Ia SYN1 0.036 synapsin I isoform Ib ZNF497 0.032 zinc finger protein 497 OBSCN 0.028 obscurin, cytoskeletal calmodulin and titin-interac... LOC100292370 0.028 PREDICTED: hypothetical protein LOC100292909 0.028 PREDICTED: hypothetical protein MYO9B 0.028 myosin IXB isoform 2 MYO9B 0.028 myosin IXB isoform 1 LOC100290946 0.028 PREDICTED: hypothetical protein XP_002347121 LOC100288840 0.028 PREDICTED: hypothetical protein XP_002344246 LOC100291774 0.028 PREDICTED: hypothetical protein XP_002346356 LOC100290694 0.028 PREDICTED: hypothetical protein XP_002348213 LOC100289421 0.028 PREDICTED: hypothetical protein XP_002343872 ARID5A 0.028 AT rich interactive domain 5A ONECUT3 0.028 one cut homeobox 3 PRAGMIN 0.024 pragmin FAM53A 0.024 dorsal neural-tube nuclear protein LOC147670 0.024 PREDICTED: hypothetical protein LOC147670 LOC147670 0.024 PREDICTED: hypothetical protein LOC147670 LOC147670 0.024 PREDICTED: hypothetical protein LOC147670 PRR12 0.024 proline rich 12 LOC100292124 0.020 PREDICTED: hypothetical protein XP_002346387Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
