
| Name: LOC100292276 | Sequence: fasta or formatted (185aa) | NCBI GI: 239754303 | |
|
Description: PREDICTED: hypothetical protein
| Not currently referenced in the text | ||
|
Composition:
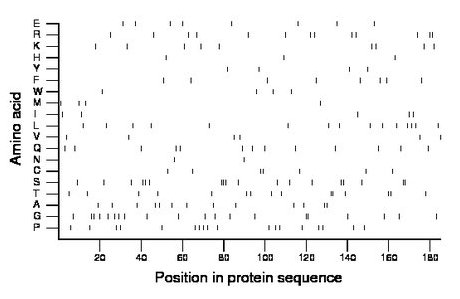
Amino acid Percentage Count Longest homopolymer A alanine 7.0 13 2 C cysteine 3.8 7 2 D aspartate 0.0 0 0 E glutamate 4.3 8 1 F phenylalanine 4.3 8 1 G glycine 10.8 20 2 H histidine 1.6 3 1 I isoleucine 2.7 5 1 K lysine 4.9 9 1 L leucine 8.1 15 1 M methionine 2.2 4 1 N asparagine 1.1 2 1 P proline 9.7 18 1 Q glutamine 7.0 13 1 R arginine 7.0 13 2 S serine 9.7 18 3 T threonine 8.1 15 2 V valine 3.2 6 1 W tryptophan 2.2 4 1 Y tyrosine 2.2 4 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein LOC100291133 1.000 PREDICTED: hypothetical protein XP_002346816 LOC100287080 1.000 PREDICTED: hypothetical protein XP_002342635 DCHS1 0.038 dachsous 1 precursor C1orf229 0.030 hypothetical protein LOC388759 EMID1 0.025 EMI domain containing 1 ITPKA 0.016 1D-myo-inositol-trisphosphate 3-kinase A LOC100127899 0.014 PREDICTED: hypothetical protein LOC727967 0.014 PREDICTED: similar to block of proliferation 1 [Hom... BOP1 0.014 block of proliferation 1 WTIP 0.014 Wilms tumor 1 interacting protein VWA5B2 0.011 von Willebrand factor A domain containing 5B2 [Homo... LOC100291048 0.011 PREDICTED: hypothetical protein XP_002347845 ZIC5 0.011 zinc finger protein of the cerebellum 5 KIAA1522 0.011 hypothetical protein LOC57648 TTC9B 0.011 tetratricopeptide repeat domain 9B BAHD1 0.011 bromo adjacent homology domain containing 1 PLCH2 0.008 phospholipase C, eta 2 RLTPR 0.008 RGD motif, leucine rich repeats, tropomodulin domain... PTCHD3 0.008 patched domain containing 3 RHPN1 0.008 rhophilin 1 LOC100293286 0.008 PREDICTED: hypothetical protein LOC100290832 0.008 PREDICTED: hypothetical protein XP_002347902 LOC100289128 0.008 PREDICTED: hypothetical protein XP_002343651 GPR98 0.008 G protein-coupled receptor 98 precursor FAM22F 0.008 hypothetical protein LOC54754 IGSF9B 0.008 immunoglobulin superfamily, member 9B RARA 0.008 retinoic acid receptor, alpha isoform 1 RARA 0.008 retinoic acid receptor, alpha isoform 1 SUPT5H 0.008 suppressor of Ty 5 homolog isoform bHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
