
| Name: LOC100292331 | Sequence: fasta or formatted (93aa) | NCBI GI: 239753621 | |
|
Description: PREDICTED: hypothetical protein
| Not currently referenced in the text | ||
|
Composition:
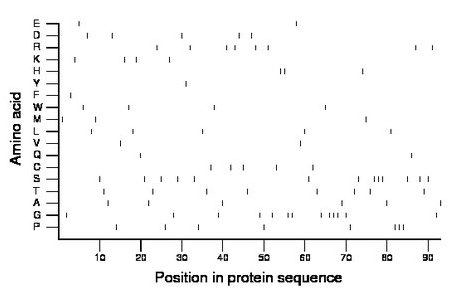
Amino acid Percentage Count Longest homopolymer A alanine 6.5 6 1 C cysteine 5.4 5 1 D aspartate 5.4 5 1 E glutamate 2.2 2 1 F phenylalanine 1.1 1 1 G glycine 14.0 13 3 H histidine 3.2 3 2 I isoleucine 0.0 0 0 K lysine 4.3 4 1 L leucine 5.4 5 1 M methionine 3.2 3 1 N asparagine 0.0 0 0 P proline 8.6 8 3 Q glutamine 2.2 2 1 R arginine 8.6 8 1 S serine 14.0 13 3 T threonine 8.6 8 1 V valine 2.2 2 1 W tryptophan 4.3 4 1 Y tyrosine 1.1 1 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein LOC100127891 0.027 PREDICTED: hypothetical protein LOC100127891 0.027 PREDICTED: similar to hCG2042508 LOC100127891 0.027 PREDICTED: similar to hCG2042508 LOC100127891 0.027 PREDICTED: similar to hCG2042508 SOX7 0.027 SRY-box 7 SETDB2 0.022 SET domain, bifurcated 2 isoform b SETDB2 0.022 SET domain, bifurcated 2 isoform a HTRA2 0.022 HtrA serine peptidase 2 isoform 1 preproprotein [Homo... HTRA2 0.022 HtrA serine peptidase 2 isoform 2 LOC100291634 0.022 PREDICTED: hypothetical protein XP_002346062 LOC100287232 0.022 PREDICTED: hypothetical protein XP_002342816 LOC100287232 0.022 PREDICTED: hypothetical protein LOC100292541 0.016 PREDICTED: hypothetical protein GATAD1 0.016 GATA zinc finger domain containing 1 GBX1 0.016 gastrulation brain homeo box 1 DLX2 0.016 distal-less homeobox 2 PKD1 0.016 polycystin 1 isoform 2 precursor PKD1 0.016 polycystin 1 isoform 1 precursor LOC100293044 0.016 PREDICTED: hypothetical protein LOC100294424 0.016 PREDICTED: hypothetical protein C4orf8 0.016 hypothetical protein LOC8603 CSPG5 0.011 chondroitin sulfate proteoglycan 5 (neuroglycan C) ... KRTAP6-3 0.011 keratin associated protein 6-3 RLTPR 0.011 RGD motif, leucine rich repeats, tropomodulin domain... FLJ37078 0.011 hypothetical protein LOC222183 DUSP8 0.011 dual specificity phosphatase 8 LOC100292758 0.011 PREDICTED: hypothetical protein NKX3-2 0.011 NK3 homeobox 2 STAB2 0.005 stabilin 2 precursorHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
