
| Name: LOC100289713 | Sequence: fasta or formatted (211aa) | NCBI GI: 239751987 | |
|
Description: PREDICTED: hypothetical protein XP_002348091
| Not currently referenced in the text | ||
|
Composition:
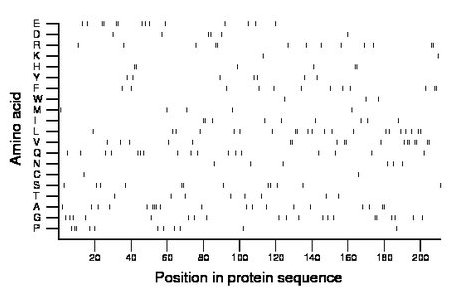
Amino acid Percentage Count Longest homopolymer A alanine 9.0 19 3 C cysteine 0.9 2 1 D aspartate 2.8 6 2 E glutamate 6.6 14 2 F phenylalanine 5.7 12 2 G glycine 9.5 20 2 H histidine 2.8 6 2 I isoleucine 4.3 9 2 K lysine 0.9 2 1 L leucine 10.0 21 2 M methionine 2.4 5 1 N asparagine 2.8 6 1 P proline 5.2 11 2 Q glutamine 8.1 17 2 R arginine 6.2 13 2 S serine 5.7 12 2 T threonine 3.8 8 1 V valine 8.5 18 2 W tryptophan 1.4 3 1 Y tyrosine 3.3 7 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein XP_002348091 BAK1 0.975 BCL2-antagonist/killer 1 BCL2 0.112 B-cell lymphoma protein 2 beta isoform BCL2L1 0.109 BCL2-like 1 isoform 1 BCL2 0.107 B-cell lymphoma protein 2 alpha isoform BCL2L2 0.097 BCL2-like 2 protein BAX 0.079 BCL2-associated X protein isoform beta BAX 0.077 BCL2-associated X protein isoform alpha BAX 0.074 BCL2-associated X protein isoform sigma MCL1 0.062 myeloid cell leukemia sequence 1 isoform 1 BCL2A1 0.055 BCL2-related protein A1 isoform 2 BCL2A1 0.055 BCL2-related protein A1 isoform 1 BOK 0.055 BCL2-related ovarian killer BAX 0.052 BCL2-associated X protein isoform delta NCOR2 0.015 nuclear receptor co-repressor 2 isoform 2 NCOR2 0.015 nuclear receptor co-repressor 2 isoform 1 MICALL1 0.015 molecule interacting with Rab13 NCOR1 0.012 nuclear receptor co-repressor 1 KIFC2 0.012 kinesin family member C2 SLC28A3 0.010 concentrative Na+-nucleoside cotransporter ARHGAP30 0.010 Rho GTPase activating protein 30 isoform 2 NUP98 0.010 nucleoporin 98kD isoform 4 NUP98 0.010 nucleoporin 98kD isoform 1 AATK 0.010 apoptosis-associated tyrosine kinase ZFP36L2 0.010 zinc finger protein 36, C3H type-like 2 TTF2 0.010 transcription termination factor, RNA polymerase II ... LRRC16B 0.007 leucine rich repeat containing 16B DKK3 0.007 dickkopf homolog 3 precursor DKK3 0.007 dickkopf homolog 3 precursor DKK3 0.007 dickkopf homolog 3 precursorHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
