
| Name: TMEM134 | Sequence: fasta or formatted (180aa) | NCBI GI: 118403326 | |
|
Description: transmembrane protein 134 isoform b
| Not currently referenced in the text | ||
Other entries for this name:
alt prot {186aa} transmembrane protein 134 isoform c alt prot {195aa} transmembrane protein 134 isoform a | |||
|
Composition:
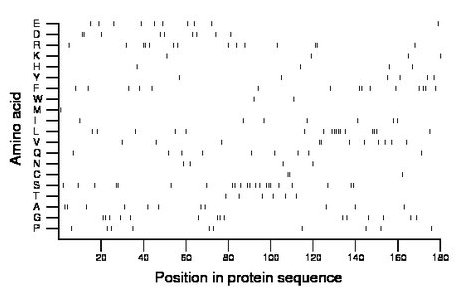
Amino acid Percentage Count Longest homopolymer A alanine 6.1 11 2 C cysteine 1.7 3 2 D aspartate 5.0 9 2 E glutamate 5.6 10 1 F phenylalanine 8.3 15 2 G glycine 8.3 15 2 H histidine 2.2 4 1 I isoleucine 3.9 7 1 K lysine 2.2 4 1 L leucine 9.4 17 5 M methionine 0.6 1 1 N asparagine 2.2 4 1 P proline 5.6 10 1 Q glutamine 5.0 9 1 R arginine 7.8 14 2 S serine 12.2 22 3 T threonine 3.3 6 1 V valine 6.1 11 2 W tryptophan 1.1 2 1 Y tyrosine 3.3 6 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 transmembrane protein 134 isoform b TMEM134 0.971 transmembrane protein 134 isoform a TMEM134 0.901 transmembrane protein 134 isoform c C20orf30 0.035 hypothetical protein LOC29058 isoform 1 C20orf30 0.035 hypothetical protein LOC29058 isoform 2 C20orf30 0.035 hypothetical protein LOC29058 isoform 2 C20orf30 0.035 hypothetical protein LOC29058 isoform 2 METRN 0.020 meteorin, glial cell differentiation regulator [Homo... LOC642423 0.018 PREDICTED: similar to hCG2004312 LOC727751 0.018 PREDICTED: similar to cis-Golgi matrix protein GM13... EPB41 0.015 erythrocyte membrane protein band 4.1 (elliptocytosi... BAHCC1 0.012 BAH domain and coiled-coil containing 1 LSM14A 0.009 LSM14 homolog A isoform a LSM14A 0.009 LSM14 homolog A isoform b PKDREJ 0.009 receptor for egg jelly-like protein precursor BRD1 0.009 bromodomain containing protein 1 SLC4A7 0.009 solute carrier family 4, sodium bicarbonate cotrans... ZBTB8A 0.009 zinc finger and BTB domain containing 8A CASS4 0.009 HEF-like protein FBXO18 0.009 F-box only protein, helicase, 18 isoform 1 FBXO18 0.009 F-box only protein, helicase, 18 isoform 2 ARL6IP4 0.009 SRp25 nuclear protein isoform 1 LOC100293005 0.006 PREDICTED: hypothetical protein LOC100289780 0.006 PREDICTED: hypothetical protein XP_002347375 LOC100289202 0.006 PREDICTED: hypothetical protein XP_002343213 PTCHD2 0.006 patched domain containing 2 LOC100293384 0.006 PREDICTED: hypothetical protein LOC100289983 0.006 PREDICTED: hypothetical protein XP_002347407 LOC100288706 0.006 PREDICTED: hypothetical protein XP_002343233 ARL6IP4 0.006 SRp25 nuclear protein isoform 3Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
